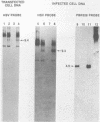
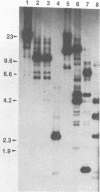
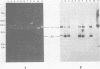
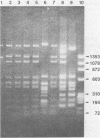
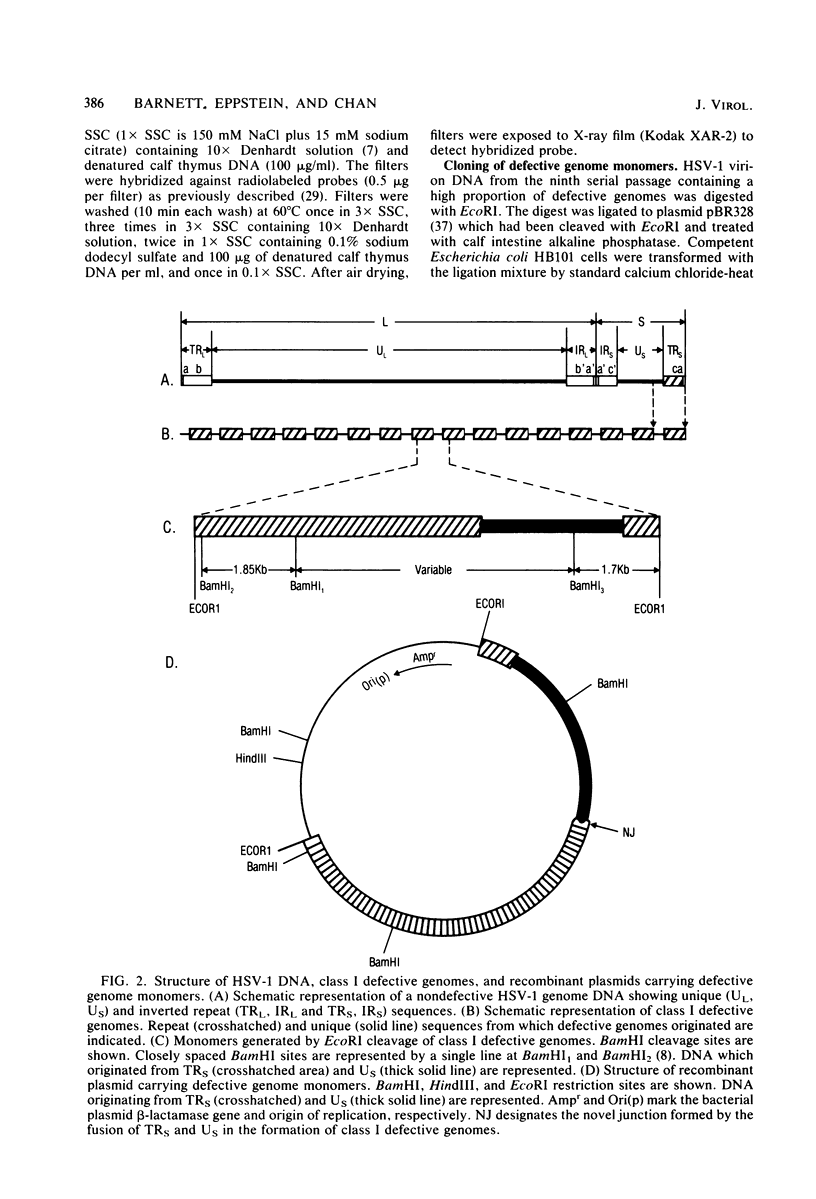
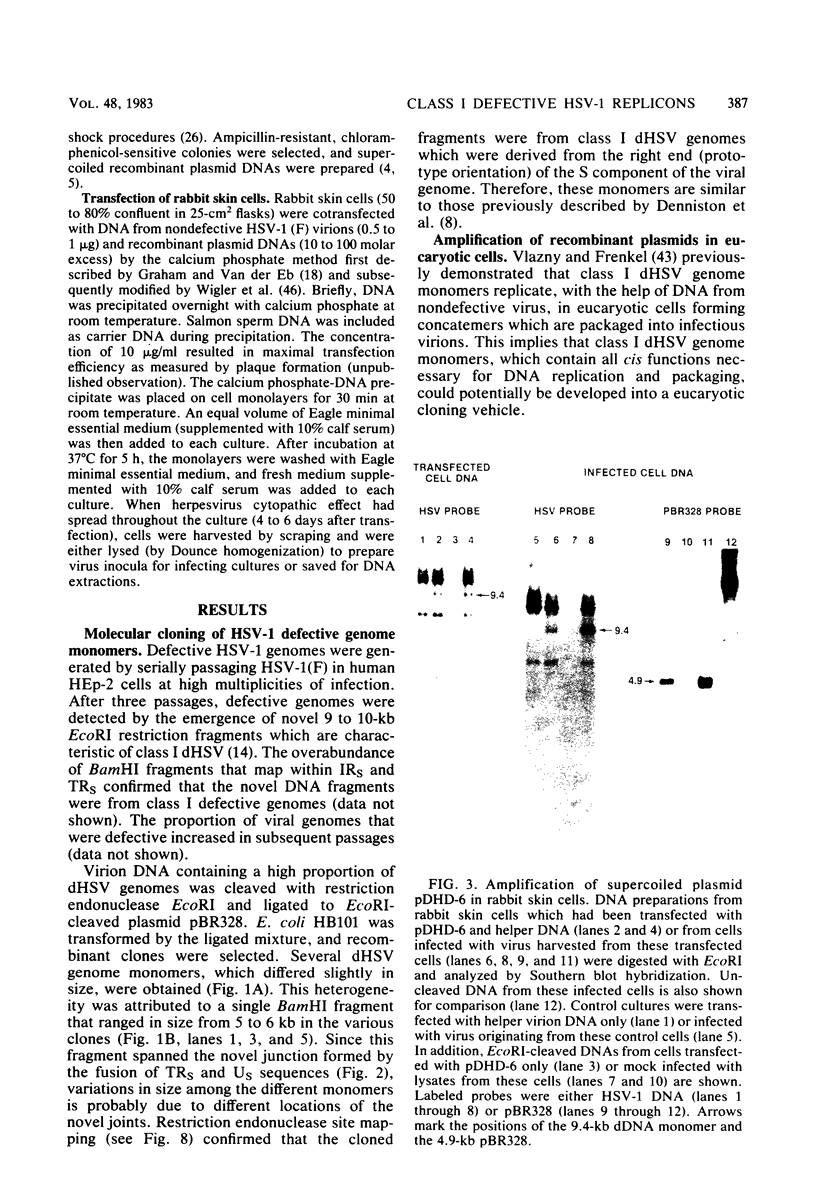
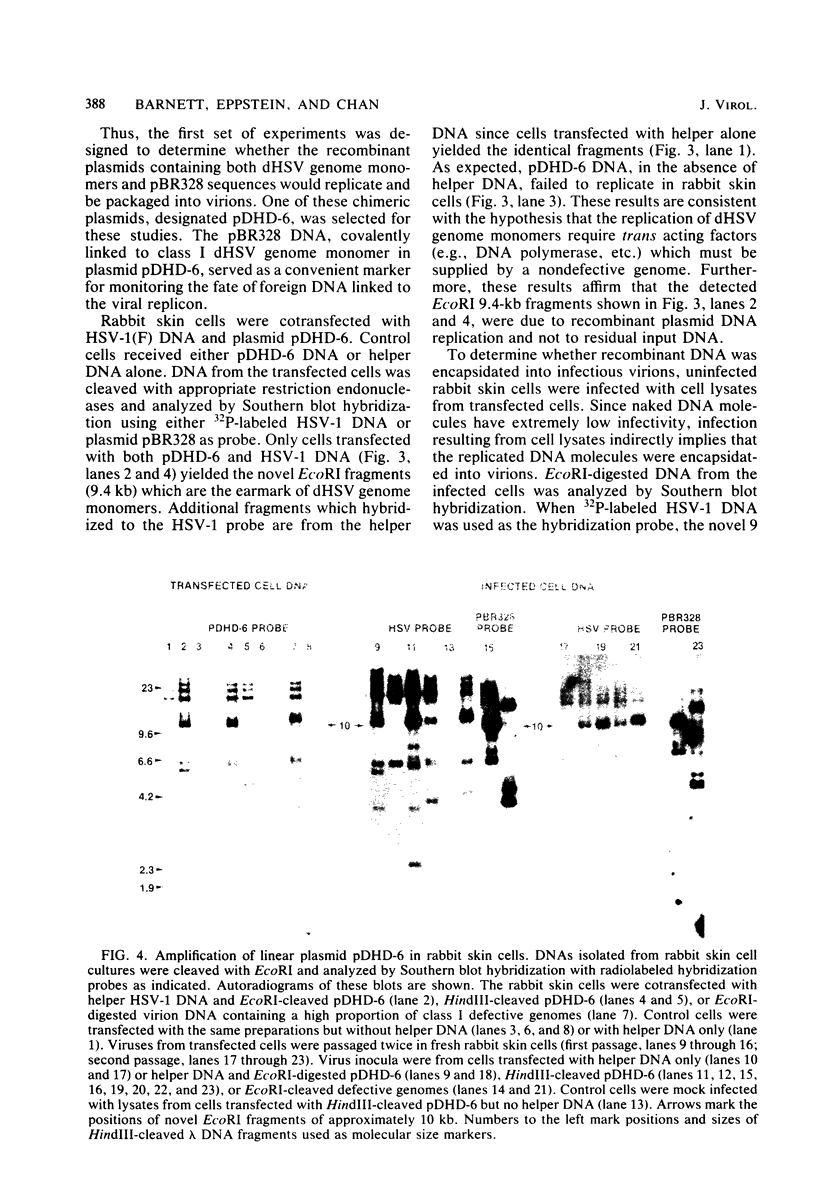
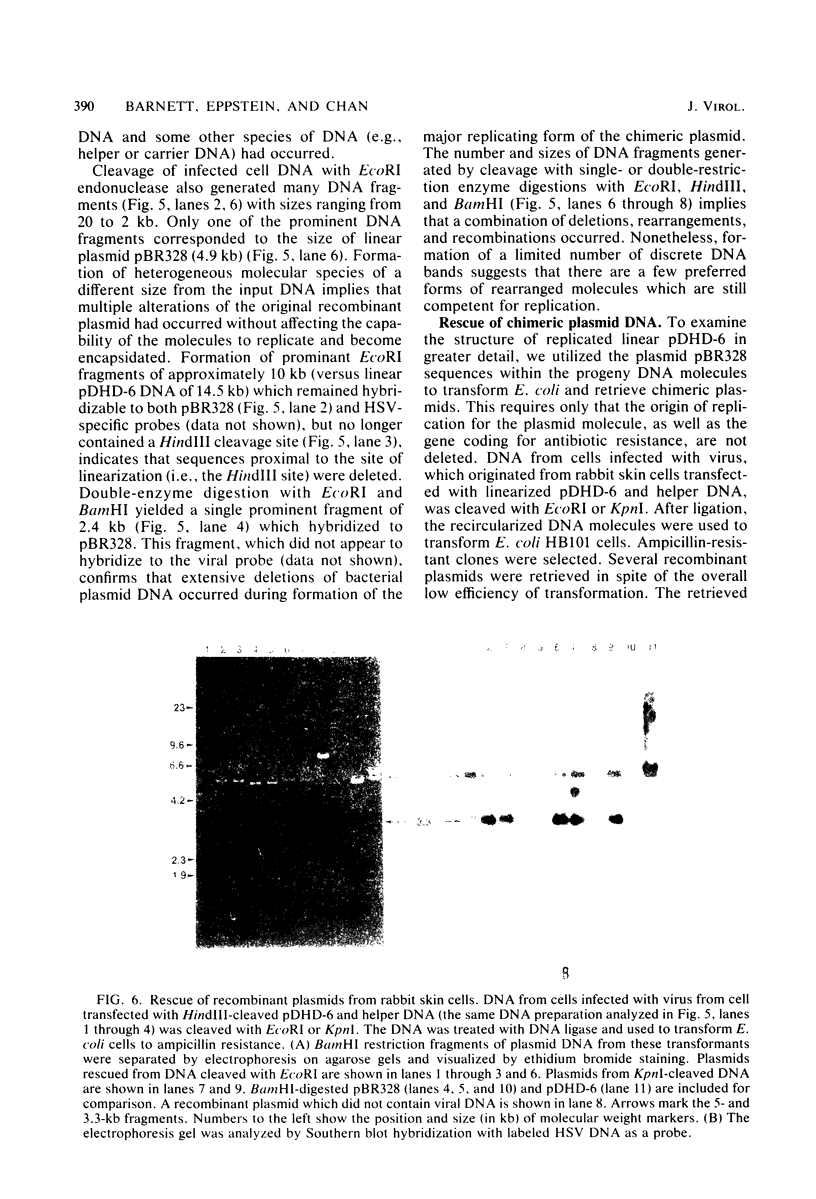
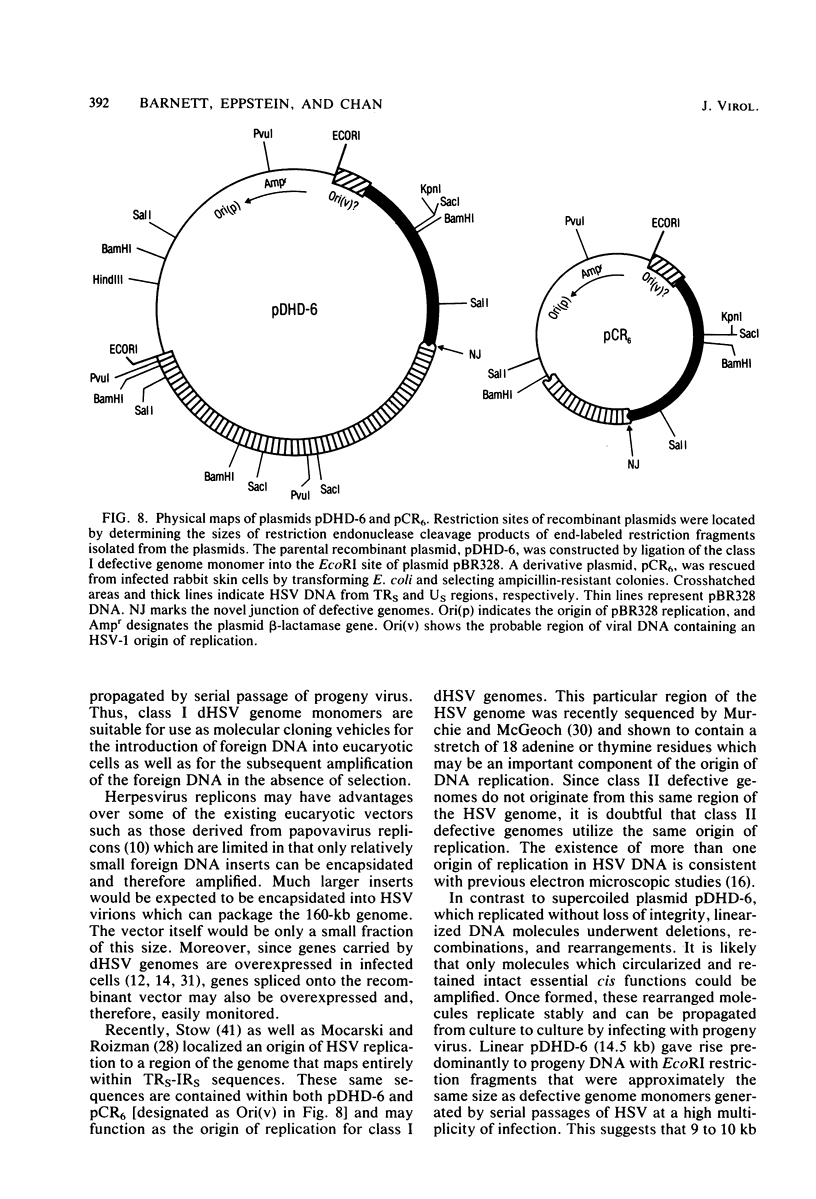
Abstract
Defective herpes simplex virus type 1 genomes are composed of head-to-tail tandem repeats of small regions of the nondefective genome. Monomeric repeat units of class I defective herpes simplex virus genomes were cloned into bacterial plasmids. The repeat units functioned as replicons since both viral and convalently linked bacterial plasmid DNA replicated (with the help of DNA from nondefective virus) when transfected into rabbit skin cells. Recombinant plasmids were packaged into virions and were propagated from culture to culture by infection with progeny virus. Replication was evidently by a rolling circle mechanism since plasmid DNA was present in a high-molecular-weight form in transfected cells. Circular recombinant plasmid DNA replicated with a high degree of fidelity. In contrast, linear plasmid DNA underwent extensive deletions of both viral and bacterial sequences when transfected into rabbit skin cells. Derivative plasmids, a fraction of the size of the parental plasmid, were rescued by transforming Escherichia coli with DNA from the transfected rabbit skin cells. These plasmids functioned as shuttle vectors since they replicated faithfully in both eucaryotic and procaryotic cells.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker Y., Asher Y., Weinberg-Zahlering E., Rabkin S., Friedmann A., Kessler E. Defective herpes simplex virus DNA: circular and circular-linear molecules resembling rolling circles. J Gen Virol. 1978 Aug;40(2):319–335. doi: 10.1099/0022-1317-40-2-319. [DOI] [PubMed] [Google Scholar]
- Bookout J., Hirsch I., Purifoy D. J., Biswal N. Herpes simplex virus types 1 and 2: comparison of the defective genomes and virus-specific polypeptides. Virology. 1979 Mar;93(2):598–604. doi: 10.1016/0042-6822(79)90265-4. [DOI] [PubMed] [Google Scholar]
- Bronson D. L., Dreesman G. R., Biswal N., Benyesh-Melnick M. Defective virions of herpes simplex viruses. Intervirology. 1973;1(3):141–153. doi: 10.1159/000148841. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Denniston K. J., Madden M. J., Enquist L. W., Vande Woude G. Characterization of coliphage lambda hybrids carrying DNA fragments from Herpes simplex virus type 1 defective interfering particles. Gene. 1981 Dec;15(4):365–378. doi: 10.1016/0378-1119(81)90180-3. [DOI] [PubMed] [Google Scholar]
- Ejercito P. M., Kieff E. D., Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968 May;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- Elder J. T., Spritz R. A., Weissman S. M. Simian virus 40 as a eukaryotic cloning vehicle. Annu Rev Genet. 1981;15:295–340. doi: 10.1146/annurev.ge.15.120181.001455. [DOI] [PubMed] [Google Scholar]
- Enquist L. W., Madden M. J., Schiop-Stanley P., Vande Woude G. F. Cloning of herpes simplex type 1 DNA fragments in a bacteriophage lambda vector. Science. 1979 Feb 9;203(4380):541–544. doi: 10.1126/science.216076. [DOI] [PubMed] [Google Scholar]
- Frenkel N., Jacob R. J., Honess R. W., Hayward G. S., Locker H., Roizman B. Anatomy of herpes simplex virus DNA. III. Characterization of defective DNA molecules and biological properties of virus populations containing them. J Virol. 1975 Jul;16(1):153–167. doi: 10.1128/jvi.16.1.153-167.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel N., Locker H., Vlazny D. A. Studies of defective herpes simplex viruses. Ann N Y Acad Sci. 1980;354:347–370. doi: 10.1111/j.1749-6632.1980.tb27977.x. [DOI] [PubMed] [Google Scholar]
- Frenkeĺ N., Locker H., Batterson W., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA. VI. Defective DNA originates from the S component. J Virol. 1976 Nov;20(2):527–531. doi: 10.1128/jvi.20.2.527-531.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann A., Shlomai J., Becker Y. Electron microscopy of herpes simplex virus DNA molecules isolated from infected cells by centrifugation in CsCl density gradients. J Gen Virol. 1977 Mar;34(3):507–522. doi: 10.1099/0022-1317-34-3-507. [DOI] [PubMed] [Google Scholar]
- Graham B. J., Bengali Z., Vande Woude G. F. Physical map of the origin of defective DNA in herpes simplex virus type 1 DNA. J Virol. 1978 Mar;25(3):878–887. doi: 10.1128/jvi.25.3.878-887.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Israel M. A., Chan H. W., Rowe W. P., Martin M. A. Molecular cloning of polyoma virus DNA in Escherichia coli: plasmid vector system. Science. 1979 Mar 2;203(4383):883–887. doi: 10.1126/science.217087. [DOI] [PubMed] [Google Scholar]
- Israel M. A., Vanderryn D. F., Meltzer M. L., Martin M. A. Characterization of polyoma viral DNA sequences in polyoma-induced hamster tumor cell lines. J Biol Chem. 1980 Apr 25;255(8):3798–3805. [PubMed] [Google Scholar]
- Jeffreys A. J., Flavell R. A. A physical map of the DNA regions flanking the rabbit beta-globin gene. Cell. 1977 Oct;12(2):429–439. doi: 10.1016/0092-8674(77)90119-2. [DOI] [PubMed] [Google Scholar]
- Kaerner H. C., Maichle I. B., Ott A., Schröder C. H. Origin of two different classes of defective HSV-1 Angelotti DNA. Nucleic Acids Res. 1979 Apr;6(4):1467–1478. doi: 10.1093/nar/6.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaerner H. C., Ott-Hartmann A., Schatten R., Schröder C. H., Gray C. P. Amplification of a short nucleotide sequence in the repeat units of defective herpes simplex virus type 1 Angelotti DNA. J Virol. 1981 Jul;39(1):75–81. doi: 10.1128/jvi.39.1.75-81.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E. D., Bachenheimer S. L., Roizman B. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J Virol. 1971 Aug;8(2):125–132. doi: 10.1128/jvi.8.2.125-132.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker H., Frenkel N. Structure and origin of defective genomes contained in serially passaged herpes simplex virus type 1 (Justin). J Virol. 1979 Mar;29(3):1065–1077. doi: 10.1128/jvi.29.3.1065-1077.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. L., Chan H. W. Identification of ecotropic proviral sequences in high- and low-ecotropic-virus-producing mouse strains. J Virol. 1982 Sep;43(3):1038–1045. doi: 10.1128/jvi.43.3.1038-1045.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie M. J., McGeoch D. J. DNA sequence analysis of an immediate-early gene region of the herpes simplex virus type 1 genome (map coordinates 0.950 to 0.978). J Gen Virol. 1982 Sep;62(Pt 1):1–15. doi: 10.1099/0022-1317-62-1-1. [DOI] [PubMed] [Google Scholar]
- Murray B. K., Biswal N., Bookout J. B., Lanford R. E., Courtney R. J., Melnick J. L. Cyclic appearance of defective interfering particles of herpes simplex virus and the concomitant accumulation of early polypeptide VP175. Intervirology. 1975;5(3-4):173–184. doi: 10.1159/000149894. [DOI] [PubMed] [Google Scholar]
- Ott A., Föhring B., Kaerner H. C. Herpes simplex virus type 1 Angelotti and a defective viral genotype: analysis of genome structures and genetic relatedness by DNA-DNA reassociation kinetics. J Virol. 1979 Feb;29(2):423–430. doi: 10.1128/jvi.29.2.423-430.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roizman B. The organization of the herpes simplex virus genomes. Annu Rev Genet. 1979;13:25–57. doi: 10.1146/annurev.ge.13.120179.000325. [DOI] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Schröder C. H., Stegmann B., Lauppe H. F., Kaerner H. C. An unusual defective genotype derived from herpes simplex virus strain ANG. Intervirology. 1975;6(4-5):270–284. doi: 10.1159/000149481. [DOI] [PubMed] [Google Scholar]
- Soberon X., Covarrubias L., Bolivar F. Construction and characterization of new cloning vehicles. IV. Deletion derivatives of pBR322 and pBR325. Gene. 1980 May;9(3-4):287–305. doi: 10.1016/0378-1119(90)90328-o. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982 Aug;30(1):295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Stegmann B., Zentgraf H., Ott A., Schröder C. H. Synthesis and packaging of herpes simplex virus DNA in the course of virus passages at high multiplicity. Intervirology. 1978;10(4):228–240. doi: 10.1159/000148986. [DOI] [PubMed] [Google Scholar]
- Stow N. D. Localization of an origin of DNA replication within the TRS/IRS repeated region of the herpes simplex virus type 1 genome. EMBO J. 1982;1(7):863–867. doi: 10.1002/j.1460-2075.1982.tb01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlazny D. A., Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci U S A. 1981 Feb;78(2):742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlazny D. A., Kwong A., Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Skare J., Summers W. C. Analysis of DNA of defective herpes simplex virus type 1 by restriction endonuclease cleavage and nucleic acid hybridization. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):683–686. doi: 10.1101/sqb.1974.039.01.082. [DOI] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978 Jul;14(3):725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]