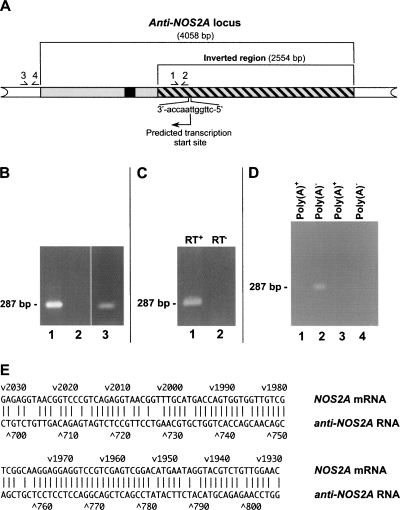
FIGURE 2.
Anti-NOS2A locus is transcribed into noncoding NOS2A-related NAT. (A) Structural organization of human anti-NOS2A locus. The inverted part of the locus is shown by the hatched box. Primers used in RT-PCR experiments are numbered. The DNA sequence of the anti-NOS2A locus has been analyzed using Promoter 2.0 software (Knudsen 1999). Notably, a region located within the inverted segment has scored 1.082, indicating the very high likelihood of the presence of a functional promoter. The promoter was predicted to drive transcription of the anti-NOS2A locus in the opposite orientation with respect to NOS2A gene. The putative transcription start site and the direction of transcription are shown by an arrow. A DNA region, transcription of which results in the synthesis of RNA fragment antisense to the NOS2A mRNA, is indicated in black. (B) The results of RT-PCR experiments performed on total RNA extracted from human meningioma (lanes 1,2) and glioblastoma (lane 3) using primers 1 and 2 (lanes 1,3) or primers 3 and 4 (lane 2). (C) The results of RT-PCR experiments performed on total RNA from human meningioma using primers 1 and 2. A sequence-specific primer complimentary to a putative RNA containing NOS2A mRNA-homologous region in antisense orientation was used in the RT reaction. Note that there is no product in the experiments in which reverse transcriptase was omitted from the first strand synthesis reaction (lane 2). (D) The results of RT-PCR experiments performed on total RNA from human meningioma using primers 1 and 2 and either poly(A)+ (lanes 1,3) or poly(A)− (lanes 2,4) RNA fractions. Note that there are no products in the experiments in which reverse transcriptase was omitted from the first-strand synthesis reactions (lanes 3,4). (E) Alignment of the antisense region of the anti-NOS2A RNA with its complementary counterpart in the NOS2A mRNA. In this alignment there is >80% complementarity.