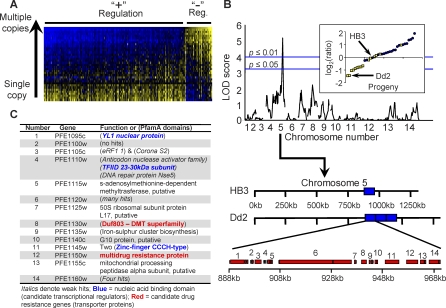
Figure 3. Transcript Levels Regulated by the pfmdr1 Amplicon on Chromosome 5.
(A) Heatmap of relative gene expression levels for 269 transcripts that mapped to the two markers corresponding to the Chr 5 amplicon containing pfmdr1 (see Figure 2, diamonds). Transcripts are aligned along the x-axis and progeny are aligned on the y-axis. The transcripts have been ranked from highest to lowest relative expression value for the Dd2 parent (top row). Progeny are ranked by their correlation to Dd2 (i.e., the bottom row is the least correlated to Dd2). Progeny that inherited a single copy of the amplicon (i.e., the HB3 parental allele) at this regulatory locus exhibited a similar expression pattern to the HB3 parent. Progeny that inherited multiple copies of the amplicon had relative expression values more similar to Dd2. Nearly all of these transcripts are distant from the amplicon (Figure 2, inset). Approximately 85% of the transcripts were positively regulated (+); inheritance of multiple copies of the regulator corresponded to increased transcription (blue). Conversely, approximately 15% of the transcripts were negatively regulated (−); inheriting multiple copies of the regulator decreased transcript levels (yellow).
(B) As an example, the eQTL mainscan for PFI0290c, a gene residing on Chr 9 whose regulation mapped to the Chr 5 amplicon. The inset illustrates the relative expression of this gene in the progeny clones; progeny inheriting the Dd2 parental allele are shown as yellow squares and those inheriting the HB3 parental allele shown as blue circles. The arrow from Chr 5 on the mainscan axis points to a schematic view of the Chr 5 amplicon and resident genes. In the Dd2, the selection for multiple copies of pfmdr1 has led to the selection of multiple copies of 13 other genes linked to pfmdr1.
(C) Nine of the 14 genes are characterized as having “unknown function” in the database. Consequently, we conducted domain searches using the PfamA database to identify possible transcriptional regulators. One gene, PFE1145w, containing two tandem zinc finger domains, characteristic of a nucleic acid binding domain common to transcription factors, was identified as a primary candidate transcriptional regulator.