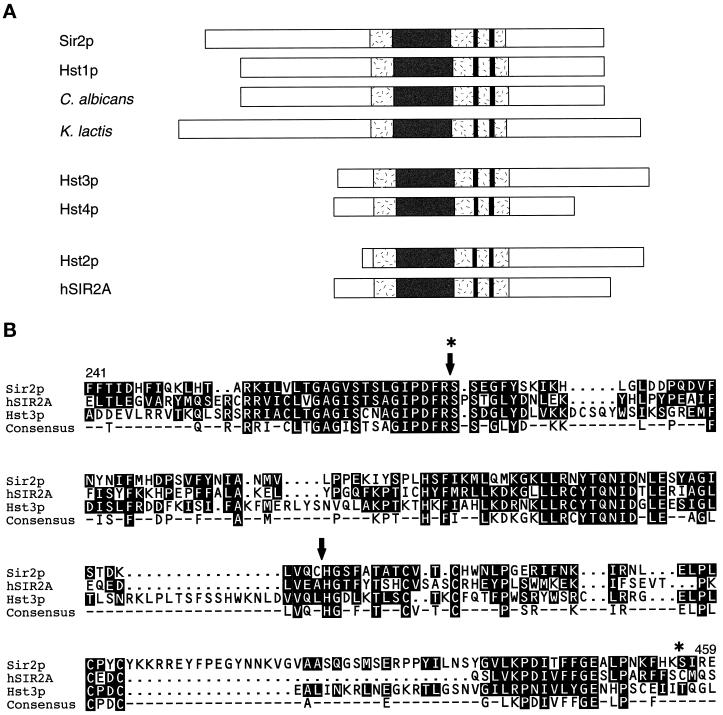
Figure 1.
Alignment of yeast Sir2p and the Hst proteins. (A) The Hst proteins from bacteria to humans are ∼30–65% identical to Sir2p overall. Sir2p and the Hst proteins all contain a characteristic core domain (shaded) that includes four cysteines of a putative zinc finger (paired lines). In the most highly conserved region of this domain (dark), bounded by the GAG and NID motifs, which are of unknown function, identity reaches 84%. The Hst proteins can be grouped into three subfamilies based on the length and sequence of their relatively distinct N and C termini. Members of the first two of these subfamilies have previously been implicated in silencing (Brachmann et al., 1995). (B) Core domain sequences of members of each of the three subfamilies (Sir2p, hSir2Ap, and Hst3p) are aligned. The consensus sequence highlights the four conserved cysteines of the putative zinc finger (at positions 372, 374, 396, and 398 in Sir2p), as well as the two conserved motifs of unknown function, which generally consist of the sequences GAG(I/V)Sxxx G(I/V)PDFRS, and (Y/I)TQNID. Some additional variation at the positions in brackets is observed for published sequences and at other positions for incomplete sequences in the databases. These motifs, although somewhat degenerate, are diagnostic of members of this gene family. The arrows and stars indicate the boundaries of the region swapped in the Sir2-hSir2A/Hst3(NID) chimeras and the Sir2-hSir2A(NID+CYS) chimera, respectively. See Brachmann et al. (1995) for additional flanking sequences.