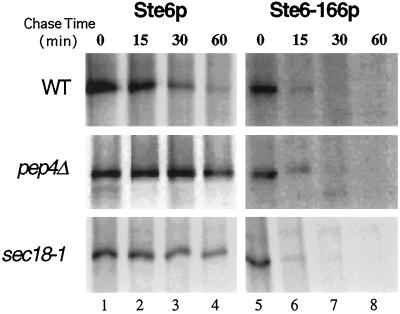
Figure 6.
The degradation of Ste6–166p is not affected in pep4▵ or sec18–1 mutants. The metabolic stability of wild-type Ste6p and Ste6–166p was examined in wild-type, pep4Δ, and sec18–1 strains. Cells were pulse-labeled for 10 min with Express35S, and the label was chased for the indicated times (min). Immunoprecipitations, detection, and quantification of the bands were performed as described in the legend to Figure 4 and in MATERIALS AND METHODS. The labeling of the sec18–1 mutant was performed after a 40-min preincubation at restrictive temperature (37°C). The isogenic wild-type SEC18 strain was submitted to the same temperature shift and showed no significant difference in the degradation of Ste6–166p (our unpublished result). The imposition of the block was confirmed by examining CPY processing in the same extracts. The p1 (ER glycosylated) form of CPY accumulated in the sec18–1 mutant at restrictive temperature, confirming the ER-to-Golgi block. The p2 form of CPY accumulated and mature CPY was absent in the pep4Δ mutant, confirming the defect in vacuolar proteolysis (our unpublished results). Strains used were SM3663 (PEP4 [CEN STE6::HAe]), SM3664 (PEP4 [CEN ste6–166::HAe]), SM3665 (pep4Δ [CEN STE6::HAe]), SM3666 (pep4Δ [CEN ste6–166::HAe]), SM3662 (sec18–1 [CEN STE6::HAe]), and SM2807 (sec18–1 [CEN ste6–166::HAe]).