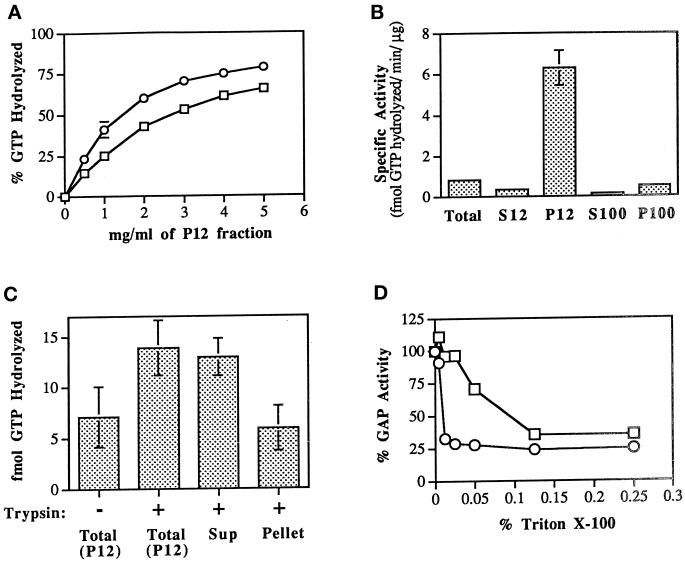
Figure 7.
Identification of a Ypt1p-GAP activity in the P12 cellular fraction and its solubilization. (A) GAP activity is linear with P12 concentration. Ypt1p (2 nM) preloaded with [γ-32P]GTP was incubated with the indicated quantities of P12 fraction at 30°C. GTP hydrolysis was measured by the charcoal-binding assay. Squares represent a 15 min incubation; circles represent a 30 min incubation. Data are expressed as the percent of the total 32P-labeled pool of GTP bound to Ypt1p that was hydrolyzed, and the intrinsic rate of GTP hydrolysis by Ypt1p was subtracted. Results are the average of two independent experiments. Error bars represent the range divided by 2. (B) Localization of GAP activity to P12 (specific activity) is shown. Crude lysates (Total) were generated from GPY60 cells. Lysates were centrifuged at 12,000 × g to generate S12 and P12. The S12 fraction was further centrifuged at 100,000 × g to generate S100 and P100. Ypt1p (2 nM; 200 fmol) preloaded with [γ-32P]GTP was incubated with the indicated cell fractions at 0.5 mg/ml for 15 min at 30°C. GTP hydrolysis was measured as described above. Specific activity is a measure of the femtomoles of GTP hydrolyzed by Ypt1p per minute per microgram of added cell fraction. Results are the average of three independent measurements performed with cell fractions from two independent fractionations. Error bars represent the SEM. (C) GAP activity is stimulated and extracted by limited trypsin digestion. Seven hundred micrograms of a P12 fraction were left untreated (Untreated Total) or treated with trypsin at 0.1 mg/ml on ice for 1 h after which time trypsin inhibitor at 0.2 mg/ml was added (Treated Total). The trypsin-treated sample was then centrifuged at 12,000 × g for 10 min to generate supernatant (Treated Sup) and pellet (Treated Pellet) fractions. The pellet was resuspended to the original volume in Buffer 88 before centrifugation. An equal volume of each sample was assayed for GAP activity as described above. Including trypsin inhibitor at the beginning of the incubation completely prevents extraction and stimulation of the GAP (our unpublished observations). Results are the average of three independent measurements. Error bars represent the SEM. (D) GAP activity is inhibited by Triton X-100. Inhibition of GAP activity in the P12 (0.5 mg/ml; squares) or in the trypsin-solubilized GAP (extracted as described in C from 0.05 mg of P12; circles) by Triton X-100 was tested. GAP assays were performed as described above in the presence of the indicated final concentrations of Triton X-100 (vol/vol) for 15 min at 30°C. Data expressed as the percent of GAP activity from the uninhibited reaction are typical of two independent experiments.