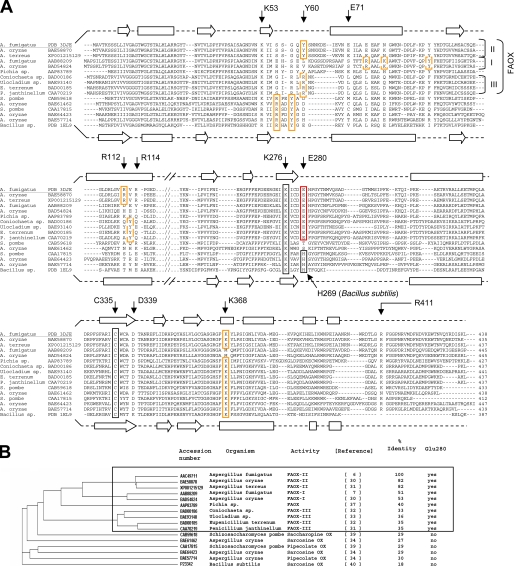
FIGURE 5.
FAOX and related proteins. A, structure-guided sequence alignment. Residues involved in carboxylate binding (Tyr-60, Arg-112, and Lys-368) in FAOX-II, MSOX and related enzymes are boxed in orange. Putative carboxylate binding residues in FAOX-I and FAOX-III are in dashed orange boxes. Residue Glu-280, involved in the binding of the fructosyl moiety, is conserved in all FAOX enzymes but not in sarcosine, pipecolate, and saccharopine oxidases(red box); Glu-280 is, therefore, a signature residue for fructosamines oxidase activity. Arg-411 also binds the fructosyl moiety and forms a conserved salt bridge together with Asp-339, which appears to be required for enzyme stability. Residues Glu-71 and Arg-114 form a salt bridge connecting the two flexible regions upon ligand binding, which is conserved only in FAOX-II. Cys-335 is covalently attached to the 8α methyl group of the flavin (black box). Lys-276 makes a water-mediated hydrogen bond with N5 of the flavin (black box). Secondary structure elements for the FAOX-II and MSOX structures are shown as arrows (β-strands) and rectangles (α-helices). B, phylogenic tree of the sequence alignment. Sequence identity is related to FAOX-II from A. fumigatus.