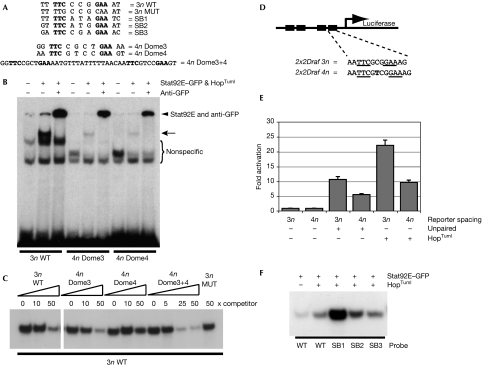
Figure 3.
Drosophila Stat binds to 3n and 4n sites with various affinities. (A) Sequences of the oligonucleotides used in (B,C,F). SB1, SB2 and SB3 correspond to the potential Stat-binding sites present in the SB fragment (Fig 1A), and Dome3, Dome4 and Dome3+4 correspond to the D. melanogaster Stat-binding sites in SLF4 (Fig 1A,B). 3n wild type (WT) corresponds to the Stat92E-binding site consensus (Yan et al, 1996). (B) EMSA assay using the radiolabelled binding sites indicated and showing binding activities that are detectable after co-transfection of plasmids expressing Stat92E–GFP and the constitutively active HopTuml proteins. Specific shifted bands corresponding to DNA:Stat92E–GFP (arrow) and DNA:Stat92E–GFP complexes supershifted with anti-GFP (arrowhead) are indicated. (C) EMSA assay using radiolabelled 3n WT to detect activated Stat92E–GFP-binding activity. Each lane contains the same quantity of cell extract and labelled 3n WT-binding site and was co-incubated with the indicated fold excess of unlabelled competitor sites. (D) Schematic representation of the reporters used in (E) with black boxes representing Stat-binding sites. Underlined bases represent the core Stat92E-binding sequence. Another thymine residue (bold) was inserted into each binding site of 2x2Draf 3n to create the 4n reporter. (E) Firefly luciferase activity in cells transfected with the reporters shown in (D) and co-transfected with plasmids expressing either the pathway ligand unpaired or the constitutively active Jak HopTuml. Levels were normalized to a co-transfected constitutively expressed Renilla luciferase plasmid and are expressed as fold change over unstimulated state. (F) EMSA assay of unactivated and activated Stat92E–GFP binding to radiolabelled WT, SB1, SB2 and SB3 oligonucleotides. EMSA, electrophoretic mobility shift assay; GFP, green fluorescent protein; MUT, mutated; STAT, signal transducer and activator of transcription.