Abstract
This study was undertaken to identify cellular proteins that bind an orally active natural product insulin mimic. Phage display cloning was used with a biotinylated derivative of this molecule as bait. Among the proteins identified was glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which has recently been shown to affect insulin receptor signaling. Binding data support a role for human GAPDH as another target of the insulin mimic, which could explain its action as a selective insulin receptor modulator.
The cytoplasmic tyrosine kinase domain of the β-chain of the insulin receptor was initially demonstrated to be the target of the small molecule insulin mimic demethylasterriquinone B1 (DAQ B1; Fig. 1).1 Identification of its cellular targets other than the insulin receptor was viewed to be useful in developing selective agents that act only on the insulin receptor and not on other proteins. Knowledge of all proteins with which DAQ B1 interacts is thus essential to evaluate the selectivity of second-generation compounds. The significance of this work derives from the aim to eventually replace injections of the hormone insulin in diabetes therapies with an orally administered drug.
Figure 1.
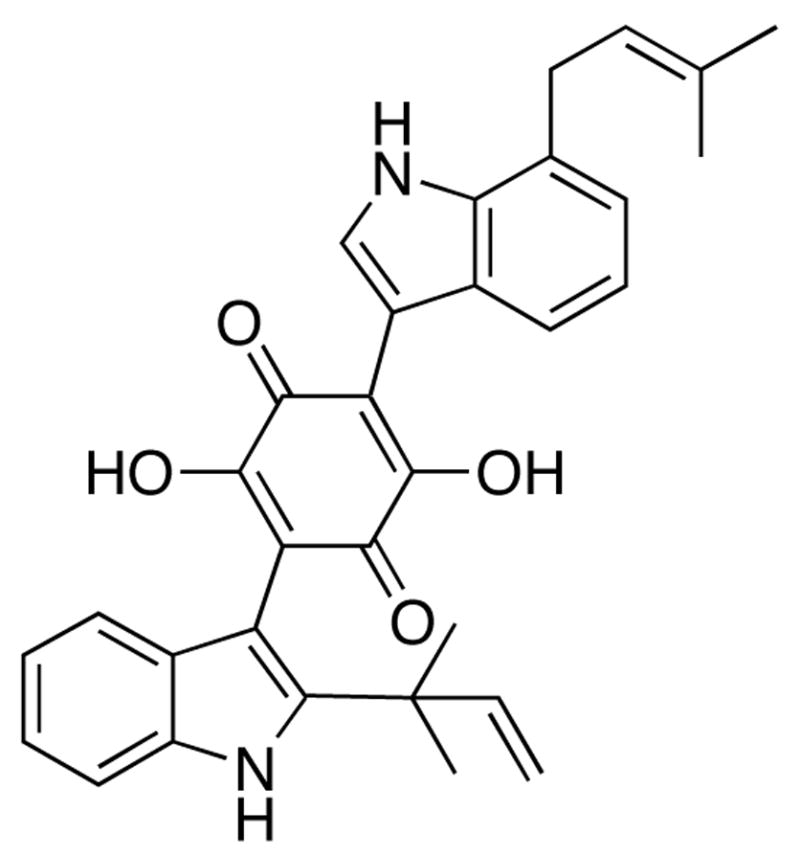
The small molecule insulin mimic demethylasterriquinone B1.
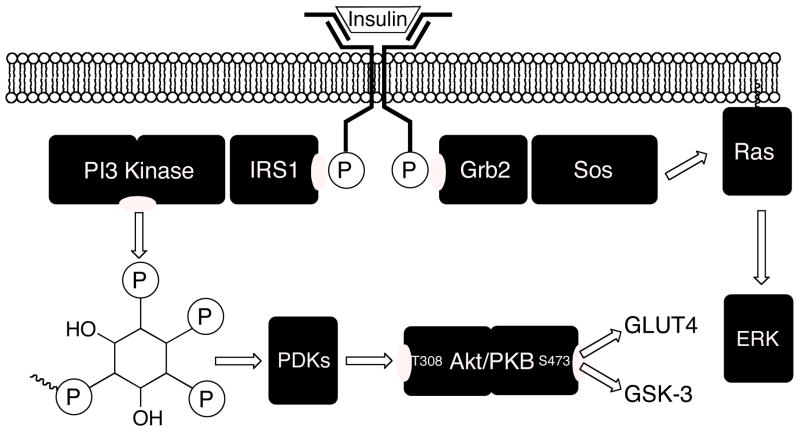
Intracellular signaling by activated insulin receptor entails two main branches (Fig. 2). One branch signals through phosphatidylinositol-3-kinase to the Akt kinase, whose phosphorylation state governs glucose transport via GLUT4 and glycogen synthesis via the GSK-3 kinase. Another branch signals through the Ras/MAPKinase (mitogen-activated protein kinase) pathway, whose control of gene transcription is related to the phosphorylation state of the ERK kinase (extra-cellular regulated kinase). This paradigm provides a useful framework for understanding some of the biological properties of DAQ B1 and its relatives. For example, DAQ B1 possesses the desirable property that, while mimicking the action of insulin in glucose lowering, it does not promote the undesired proliferation of vascular smooth muscle cells of the endothelium, as occurs in insulin resistance2,3 and with insulin administration.4 Investigations of the basis for this effect suggested that DAQ B1 selectively stimulates the Akt branch of insulin signal transduction at the expense of the ERK branch, whereas insulin stimulates both equally.5,6 While we hypothesized that the action of DAQ B1 on other cellular protein targets could be responsible for this observation, the mechanism by which it could accomplish this task was unexplained and the target protein(s) were unidentified. We therefore aimed to identify potential protein binding partners for DAQ B1 using phage display cloning.
Figure 2.
Insulin receptor signaling. Upon activation by extracellular ligand binding, insulin receptor first phosphorylates tyrosine residues in its cytoplasmic β-chain. One branch of the signal transduction pathway involves phosphatidylinositol-3-kinase (PI3-kinase), its product phosphatidylinositol-3,4,5-tris-phosphate, and its signaling to the Akt kinase. This branch promotes glucose uptake and glycogen synthesis. The other branch of insulin receptor signal transduction is the mitogen-activated protein kinase cascade common to several growth factor signaling pathways. It promotes gene expression and cellular proliferation.
Methods to search broadly for the protein binding partners of small molecules have been developed by Austin7,8,9 and Hidaka.10,11 Like other strategies,12 these methods link the small molecule to a solid phase to be used as an affinity matrix. However, rather than using this matrix to purify binding protein(s) from a cellular extract, which requires subsequent protein purification, sequencing, and identification, these methods instead use a library of proteins expressed on the surface of T7 phage. Phage display cloning is far easier than affinity protein purification because protein identification comes directly from the cDNA in the phage. This method has recently been applied to finding a novel binding partner for the long-known drug doxorubicin.13 Austin’s method is particularly attractive because a biotinylated form of the drug is used, reducing the synthetic task to the introduction of biotin at a position that does not interfere with biological activity. Such biotin-drug conjugates are useful for other purposes, such as studies of drug transport, identification of sub-cellular localization, or the direct determination of binding affinities.
Using the method of methyl scanning, we recently identified the sites at which DAQ B1 could be functionalized without compromising its cell-based insulin mimetic activity.14 This work also reported the preparation of a DAQ B1 analogue derivatized with biotin at one position. This compound, called DAQ-biotin (Fig. 3), was used as bait in phage display cloning using a T7 phage library. One outcome not expected from this experiment was the cloning of insulin receptor. As an integral membrane protein, insulin receptor is not expected to be properly expressed as a fusion with a T7 coat protein. However, other cellular proteins with binding affinity to DAQ B1 could be properly expressed and thereby be identified.
Figure 3.
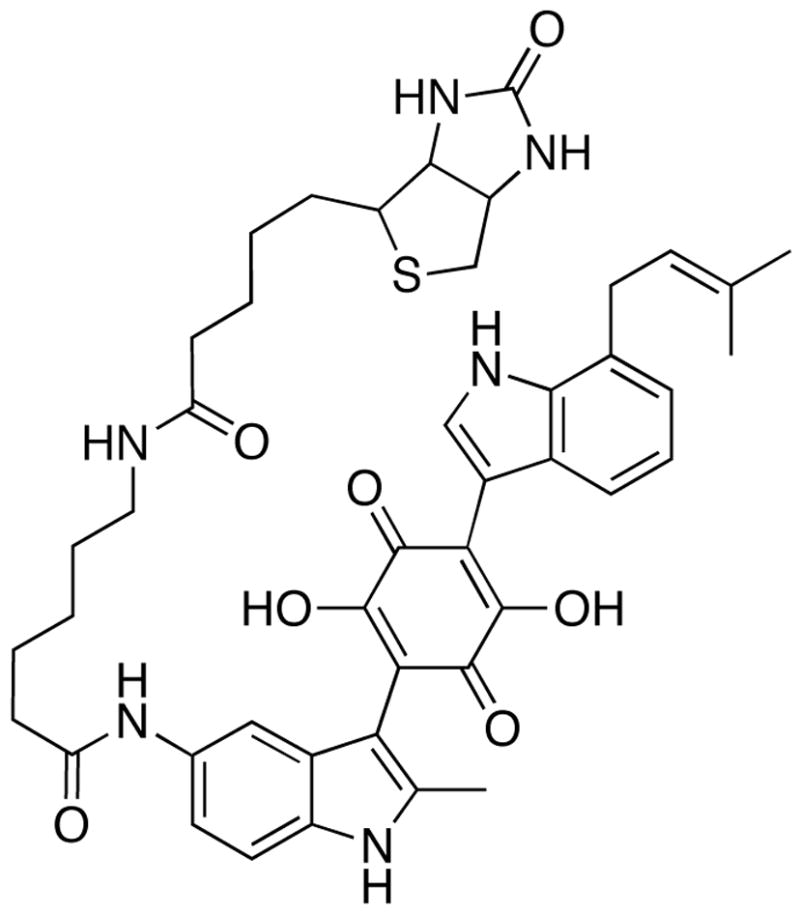
The bait molecule DAQ-biotin. Its structure preserves the major pharmacophore of DAQ B1, the 7-prenyl indole and the central quinone, while attaching biotin through a linker arm that permits its immobilization on an affinity matrix via avidin.
Biopanning of a T7 phage human liver cDNA library with DAQ-biotin was performed using protocols adapted from those of Austin and the library vendor (see Supporting Information). The drug conjugate was immobilized on a NeutrAvidin-coated 96-well plate and at least five rounds of biopanning were performed. The enrichment of phage binding to DAQ-biotin was evaluated by polymerase chain reaction (PCR) and agarose gel electrophoresis, examining the size of the cDNA inserts into the gene for the phage coat protein cp10. Biopanning continued until analysis of the resulting total phage lysate by PCR gave less than 10 different fragment sizes (500 to 1500 nt). Of course, each sized insert includes many different cDNAs. Following biopanning, phage lysates were cloned out for individual analysis. To minimize repetitive sequencing of the same cDNA, phage with the same insert sizes were further sorted by restriction mapping of the inserts using enzymes with 4-base recognition sites. The restriction patterns appearing most frequently were regarded as the phage most likely to bear DAQ B1 binding proteins, and PCR products from these clones were submitted for DNA sequencing. The sequences of these clones were analyzed by BLAST against the human genome, with the results summarized in Table 1. While explanations for some of these putative DAQ B1 binding proteins are elusive, they include fibrinogen α chain. Because angiopoietin-like 3 includes a fibrinogen domain,15 this motif may be one involved in binding to DAQ B1.
Table 1.
Proteins identified as potential DAQ B1 targets using phage display cloning, ranked by decreasing frequency of appearance of that clone
| Protein | subsequence found in the clone |
|---|---|
| NADH dehydrogenase subunit 4 | +3 ATELIIFYIFFETTLIPTLAIITRGNQPERLNAGTYFLFYTLVGSLPLLIA SKHIAYPLPWYYPYGGIIITSSICLRQTDLKSLIAYSSISHIALVVTAILIQTP 54 |
| tight junction protein 1, isoform b | LXVXPDPPSIFRISQQLSDHQTEKILRQAAFYPQKSFPDKAPVNGTEQTQKTVTPAYNRF TPKPYTSSARPFERKFESPKFNH 83 |
| angiopoietin-like 3 precursor | PGKQDLVFSTWDHKAKGHFNCPEGYSGGWWWHDECGENNLNGKYNKPRAKSKPERRRGLS WKSQNGRLYSIKSTKMLIHPTDSG 84 |
| tetratricopeptide repeat domain 14 isoform a | RDDRGXIYGYRRFEKDIEGRKEHYRRWEPGSVRHSTSPASSEYSWKSVEKYKKYAHSGSRDFSR HEQRYRLNTNQGEYEREDNYGEDIKTEVPEEDALSSKEHSESSVKK 110 |
| glyceraldehyde-3-phosphate dehydrogenase | REGGPPSRASWATLSTRVVSSDFNSDTHSSTFDAGAGIALNDHFVKLISWYDNEFGYSNR 60 VVDLMAHMASKE 72 |
| fibrinogen alpha chain | AGTGPLKSSVSGSTGQWHSESGSFRPDSPGSGNARPNNPDWGTFEEVSGNVSPGTRREYH 60 TEKLVTSKGDKELRTGKEKVTSGSTTTTRRSCSKTVTKTVIGPDGHKEVTKEVVTSEDGS 120 DCPEAMDLGTLSGIGTLDGFRHRHPDEAAFFDTASTGKTFPGFFSPMLGEFVSETESRGS 180 ESGIFTNTKESSSHHPGIAEFPSRGKSSSYSKQFTSSTSYNRGDSTFESKSYKMADEAGS 240 EADHEGTHSTKRGHAKSRPVRGIHTSPLGEAFPCP 275 |
| unknown protein | QTHSLPPSLVLSLWRHNYNKLHLPTTNRPKIAHCILFNQPHSPRSNSHSHPNPLKLAAALE 61 |
| unnamed gene product | +2 GHRTNHILYLLRNHTYPHLGYHHPMRQPARTPERRHLLPILHPSRLPSPTHRT 58 |
| unnamed gene product | APGWSPLTSTATPTPPPLTLGLALPSTTTLSSSFPGMTTNLATATGWWTSWPTWPPRSKT 60 PGPPAPARAQEEERDPHCWGVPATLSPPPH 90 |
| putative protein | +3 PLILAIIPIPPYIQTTKHNISPTKPITLLTPSRRPPHSNLNRRTTSKLPFYHHWTSSIRTILHNNPNPNT 70 |
| hypothetical primate protein | DPWTTSPSKSTRGRERPSLLGSPCHTQSPTTLNLPSSQLPCRPLEEGSGLGSRTLSCTINKVC 63 |
| similar to primate cytochrome 10 ORF 111 | QRIWLQQQGGGPHGPHGLQGVRPLDHQPQQEHKRKRETLTAGESLPHSVPHHTESPLLTVAM 62 |
Most interesting was the identification of human GAPDH. While well known as a glycolytic enzyme, GAPDH is also involved in several non-glycolytic functions (e.g. transcription, DNA repair, apoptosis, and nuclear RNA export),16 and it binds strongly with the glucose transporter.17 Identification of GAPDH in this study is especially provocative in light of the recent report from Chang et al. that GAPDH is the target of a molecule they called GAPDS that reverses the dauer phenotype in C. elegans mutants, which is caused by impaired insulin signaling.18 The biochemical mechanism by which GAPDH plays a role in insulin signaling is still unclear, but several lines of evidence support a model in which a functional GAPDH is essential for a cellular phosphatase activity that acts on phosphoinositides (such as phosphatidylinositol-3,4,5-tris-phosphate, Fig. 2). Evidence provided by Chang et al. suggests that GAPDH must be in its tetrameric form to promote this cellular lipid phosphatase activity. Therefore, impairing GAPDH tetramerization through a small molecule binding event maintains the phosphorylation state of phosphoinositides that is essential for insulin signaling to Akt. The ability of DAQ B1 to simultaneously activate insulin receptor and inhibit lipid phosphatase action upon phosphatidylinositol-3,4,5-tris-phosphate could explain its selective activation of Akt in 3T3-L1 pre-adipocytes as we earlier described.5
To confirm the binding of DAQ B1 to GAPDH and measure its affinity, surface plasmon resonance was used (Fig. 4). Assays were performed by immobilizing human erythrocyte GAPDH on a dextran-coated chip. DAQ B1 and DAQ-biotin were used as the binding analytes. Their KDs are 7.2 μM and 7.5 μM, respectively. The close correspondence of these two values shows that the attachment of biotin to the DAQ B1 structure does not affect its binding to GAPDH, as predicted by the methyl scanning study. Neither data set is an excellent fit to a pure 1:1 binding model (χ2 = 0.44), but they fit better to a 2-state, conformational change binding model (χ2 = 0.37), behavior expected for a molecule that causes dissociation of tetrameric GAPDH following binding. Further studies are needed to establish that DAQ B1 directly affects the tetrameric structure of GAPDH. However, the observed maximum binding of DAQ B1 was 4× lower than expected for a 1:1 interaction. While the measured binding affinity of DAQ B1 for human GAPDH is perhaps less than might have been expected, the small molecule identified by Chang et al. to reverse the dauer phenotype of C. elegans inhibits the enzymatic activity of rabbit muscle GAPDH with an IC50 of 15 μM. Therefore, binding affinities of this order of magnitude can still have profound biological consequences.
Figure 4.
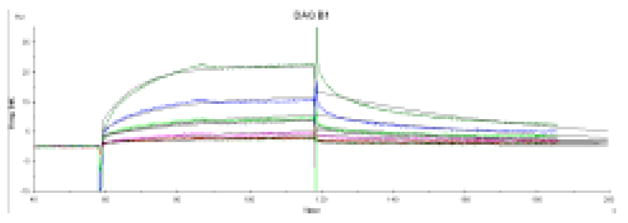
Surface plasmon resonance (SPR) data. The sensorgram of the binding of free DAQ B1 to immobilized human erythrocyte GAPDH. Analysis was conducted at concentrations from 40 μM to 2.5 μM. Binding parameters determined from fitting these data to a 2-state binding model are: kA 2.18 × 104 M−1 · s−1; kD 1.57 × 10−2 s−1; KD 7.2 μM. DAQ B1 and DAQ-biotin both behave in a manner characteristic of dose-dependent binding to GAPDH with a high level of specificity. Non-specific binding often shows SPR responses greater than theoretical, but the observed responses were less than the maximum predicted, which is explained by the expected loss of active GAPDH upon immobilization.
Phage display cloning offers the possibility of not only identifying the protein(s) that recognize(s) the small molecule, but also identifying the sub-domains responsible for binding. This is because the expressed cDNAs can be less than full-length. The region of human GAPDH identified as binding to DAQ B1 is the extreme C-terminus. X-ray structures of human GAPDHs have been determined19 and are similar to earlier structures of bacterial GAPDHs.20 The C-terminus in all GAPDHs is a mixed fold including the α3 helix and the anti-parallel β6, β7 and β8 sheets. A specific role in tetramerization has not been ascribed to any region of GAPDH.21
This study supports the idea that the action of DAQ B1 on multiple protein targets explains its action as a selective insulin receptor modulator. It is ironic that it was aimed at identifying new protein targets in order to design out their interactions with future drug candidates, but the action of DAQ B1 against one of these targets might prove to be one of its major advantages. Further biochemical studies are needed to establish the details and consequences of the interactions between DAQ B1 (and related next-generation compounds) and human GAPDH in their biological properties, specifically glucose lowering in cells without promotion of cellular proliferation.
The major finding of this work is that a cellular binding partner for demethylasterriquinone B1, an orally active small molecule activator of the human insulin receptor, is human GAPDH. This study and the C. elegans work of Chang et al. strengthen one another because they independently converge on the same target, GAPDH. Chang concluded that this enzyme in its tetrameric form is essential for a cellular phosphatase activity that dephosphorylates phosphatidylinositol lipids produced by phosphatidylinositol-3-kinase (PI3Kinase). Binding of small molecules, such as demethylasterriquinone B1, to GAPDH could therefore disrupt phosphatase(s) acting upon phosphatidylinositol lipids and thereby potentiate insulin signaling via the PI3Kinase pathway. The PI3Kinase branch of the insulin signaling pathway promotes glucose transport. The MAPKinase branch of the insulin signaling pathway promotes proliferation of vascular smooth muscle cells, an important step in the atherosclerotic process. The abilities of demethylasterriquinone B1 to promote human insulin receptor phosphorylation (stimulating both pathways) and to enhance signaling in the PI3Kinase branch of intracellular insulin signal transduction by inhibiting a lipid phosphatase activity explains its previously described action as a selective insulin receptor modulator. Such molecules not only offer the potential advantage of far easier administration than the native hormone insulin, but also of more beneficial medicinal action.
The 1999 discovery of the insulin mimetic activity of DAQ B1 represented a breakthrough in small molecule replacements for human therapeutic proteins. Through extensive medicinal chemistry, a potent and selective compound related to DAQ B1 was identified and further tested in rats, dogs, and primates.22 Yet, this molecule has not advanced into clinical trials. While there could be many reasons for this, it is certainly possible that incomplete understanding of all of the cellular interactions of a drug candidate such as this one could impede its clinical development. Information on all of the proteins in a cell with which a small molecule can interact should be invaluable for the consideration of potential off-target activities and toxicity of a drug candidate. The phage display cloning method, in combination with an efficient synthetic route and the methyl scanning method, permits many potential biological targets of any small molecule to be identified. This information is gained without the need for biochemistry or protein chemistry, and only chemical synthesis and molecular biology, which are more widely available expertise, are needed. Application of this paradigm to all small molecule candidates for clinical development merits consideration.
Supplementary Material
Materials and experimental descriptions for biopanning. PCR analysis. cloning. and surface plasmon resonance (4 pages). This material is available free on the Internet at pubs.acs.org.
Acknowledgments
Financial support from the NIH (DK-60532) is gratefully acknowledged. The assistance of V. Smola in administrative support of this work is appreciated.
Footnotes
Abbreviations: DAQ B1, demethylasterriquinone B1; ERK kinase, extra-cellular regulated kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MAPKinase, mitogen-activated protein kinase; PCR, polymerase chain reaction; PI3Kinase, phosphatidylinositol-3-kinase; SPR, surface plasmon resonance.
References
- 1.Zhang B, Salituro G, Szalkowski D, Li Z, Zhang Y, Royo I, Vilella D, Diez MT, Pelaez F, Ruby C, Kendall RL, Mao X, Griffin P, Calaycay J, Zierath JR, Heck JV, Smith RG, Moller DE. Discovery of a small molecule insulin mimetic with antidiabetic activity in mice. Science. 1999;284:974–977. doi: 10.1126/science.284.5416.974. [DOI] [PubMed] [Google Scholar]
- 2.Bansilal S, Farkouh ME, Fuster V. Role of insulin resistance and hyperglycemia in the development of atherosclerosis. Am J Cardiol. 2007;99:6B–14B. doi: 10.1016/j.amjcard.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Uwaifo GI, Ratner RE. The roles of insulin resistance, hyperinsulinemia, and thiazolidinediones in cardiovascular disease. Am J Med. 2003;115(Suppl 8A):12S–19S. doi: 10.1016/j.amjmed.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 4.Weber MA, Lidor A, Arora S, Salituro GM, Zhang BB, Sidawy AN. A novel insulin mimetic without a proliferative effect on vascular smooth muscle cells. J Vasc Surg. 2000;32:1118–26. doi: 10.1067/mva.2000.111280. [DOI] [PubMed] [Google Scholar]
- 5.Webster NJG, Park K, Pirrung MC. Signaling effects of demethylasterriquinone B1: a selective insulin receptor modulator (SIRM) ChemBioChem. 2003;4:379–85. doi: 10.1002/cbic.200200468. [DOI] [PubMed] [Google Scholar]
- 6.Ding VD, Qureshi SA, Szalkowski D, Li Z, Biazzo-Ashnault DE, Xie D, Liu K, Jones AB, Moller DE, Zhang BB. Regulation of insulin signal transduction pathway by a small-molecule insulin receptor activator. Biochem J. 2002;367:301–6. doi: 10.1042/BJ20020708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sche PP, McKenzie KM, White JD, Austin DJ. Display cloning: functional identification of natural product receptors using cDNA-phage display. Chem Biol. 1999;6:707–16. doi: 10.1016/s1074-5521(00)80018-6. [DOI] [PubMed] [Google Scholar]
- 8.Savinov SN, Austin DJ. The cloning of human genes using cDNA phage display and small-molecule chemical probes. Comb Chem High Throughput Screen. 2001;4:593–7. doi: 10.2174/1386207013330814. [DOI] [PubMed] [Google Scholar]
- 9.McKenzie KM, Videlock EJ, Splittgerber U, Austin DJ. Simultaneous identification of multiple protein targets by using complementary-DNA phage display and a natural-product-mimetic probe. Angew Chem Int Ed. 2004;43:4052–5. doi: 10.1002/anie.200454004. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka H, Ohshima N, Hidaka H. Isolation of cDNAs encoding cellular drug-binding proteins using a novel expression cloning procedure: drug-western. Mol Pharmacol. 1999;55:356–63. doi: 10.1124/mol.55.2.356. [DOI] [PubMed] [Google Scholar]
- 11.Ikenoya M, Doi T, Miura T, Sawanobori K, Nishio M, Hidaka H. Investigation of binding proteins for anti-platelet agent K-134 by drug-western method. Biochem Biophys Res Commun. 2007;353:1111–4. doi: 10.1016/j.bbrc.2006.12.172. [DOI] [PubMed] [Google Scholar]
- 12.Sin N, Meng L, Auth H, Crews CM. Eponemycin analogues: syntheses and use as probes of angiogenesis. Bioorg Med Chem. 1998;6:1209–17. doi: 10.1016/s0968-0896(98)00089-3. [DOI] [PubMed] [Google Scholar]
- 13.Jin Y, Yu J, Yu YG. Identification of hNopp140 as a binding partner for doxorubicin with a phage display cloning method. Chem Biol. 2002;9:157–62. doi: 10.1016/s1074-5521(02)00096-0. [DOI] [PubMed] [Google Scholar]
- 14.Pirrung MC, Liu Y, Deng L, Halstead DK, Li Z, May JF, Wedel M, Austin DA, Webster NJG. Methyl scanning: total synthesis of demethylasterriquinone B1 and derivatives for identification of sites of interaction with and isolation of its receptor(s) J Am Chem Soc. 2005;127:4609–24. doi: 10.1021/ja044325h. [DOI] [PubMed] [Google Scholar]
- 15.Conklin D, Gilbertson D, Taft DW, Maurer MF, Whitmore TE, Smith DL, Walker KM, Chen LH, Wattler S, Nehls M, Lewis KB. Identification of a mammalian angiopoietin-related protein expressed specifically in liver. Genomics. 1999;62:477–82. doi: 10.1006/geno.1999.6041. [DOI] [PubMed] [Google Scholar]
- 16.Sirover MA. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- 17.Lachaal M, Berenski CJ, Kim J, Jung CY. An ATP-modulated specific association of glyceraldehyde-3-phosphate dehydrogenase with human erythrocyte glucose transporter. J Biol Chem. 1990;265:15449–54. [PubMed] [Google Scholar]
- 18.Min J, Kyung Kim Y, Cipriani PG, Kang M, Khersonsky SM, Walsh DP, Lee JY, Niessen S, Yates JR, 3rd, Gunsalus K, Piano F, Chang YT. Forward chemical genetic approach identifies new role for GAPDH in insulin signaling. Nat Chem Biol. 2007;3:55–9. doi: 10.1038/nchembio833. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins JL, Tanner JJ. High-resolution structure of human D-glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2006;62:290–301. doi: 10.1107/S0907444905042289. [DOI] [PubMed] [Google Scholar]; Ismail SA, Park HW. Structural analysis of human liver glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2005;61:1508–13. doi: 10.1107/S0907444905026740. [DOI] [PubMed] [Google Scholar]
- 20.Biesecker G, Harris JI, Thierry JC, Walker JE, Wonacott AJ. Sequence and structure of D-glyceraldehyde 3-phosphate dehydrogenase from Bacillus stearothermophilus. Nature. 1977;266:328–33. doi: 10.1038/266328a0. [DOI] [PubMed] [Google Scholar]
- 21.Nagradova NK. Interdomain interactions in oligomeric enzymes: creation of asymmetry in homo-oligomers and role in metabolite channeling between active centers of hetero-oligomers. FEBS Lett. 2001;487:327–32. doi: 10.1016/s0014-5793(00)02338-3. [DOI] [PubMed] [Google Scholar]
- 22.Liu K, Xu L, Szalkowski D, Li Z, Ding V, Kwei G, Huskey S, Moller DE, Heck JV, Zhang BB, Jones AB. Discovery of a potent, highly selective, and orally efficacious small-molecule activator of the insulin receptor. J Med Chem. 2000;43:3487–94. doi: 10.1021/jm000285q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and experimental descriptions for biopanning. PCR analysis. cloning. and surface plasmon resonance (4 pages). This material is available free on the Internet at pubs.acs.org.