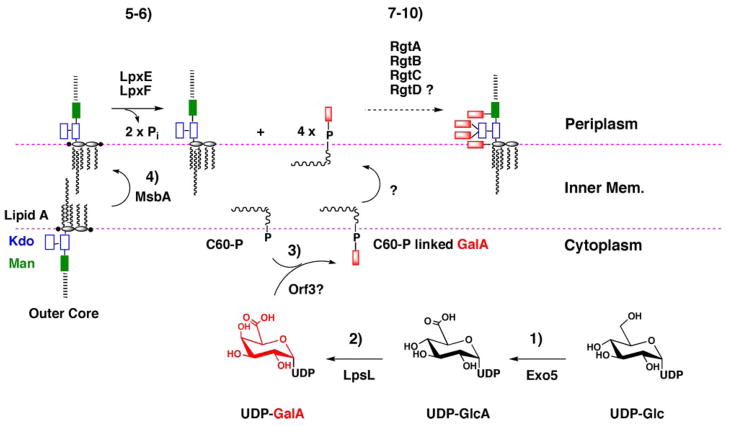
FIGURE 9. Proposed pathway and topography for attachment of GalA residues to the lipid A and core domains of R. leguminosarum LPS.
The simplest scenario for the biosynthesis of dodecaprenyl-β-D-GalA is as follows. 1 and 2, UDP-Glc is oxidized by the dehydrogenase Exo5 to UDP-GlcA, which is then converted by the C4-epimerase LpsL to UDP-GalA (22). 3, in analogy to PmrF (ArnC) (30) in E. coli, Orf3 transfers GalA from UDP-GalA to dodecaprenyl phosphate, generating dodecaprenyl phosphate-β-D-GalA, which is flipped to the periplasmic leaflet by an unknown mechanism. As yet, we have not been able to assay Orf3 in vitro. 4, lipid A and core LPS sugars are synthesized on the cytoplasmic leaflet of the inner membrane (43–46, 56, 57) and flipped to the periplasmic leaflet by MsbA. 5 and 6, on the periplasmic side of the inner membrane the 1- and 4′-phosphatases, LpxE and LpxF (40, 50), dephosphorylate lipid A, creating the substrate for GalA addition. 7–10, GalA is transferred from dodecaprenyl-β-D-GalA to the outer Kdo by RgtA and RgtB, and to mannose by RgtC. The same GalA donor is presumably used in the modification of lipid A by RgtD, but an in vitro assay is not yet available.