Figure 7.
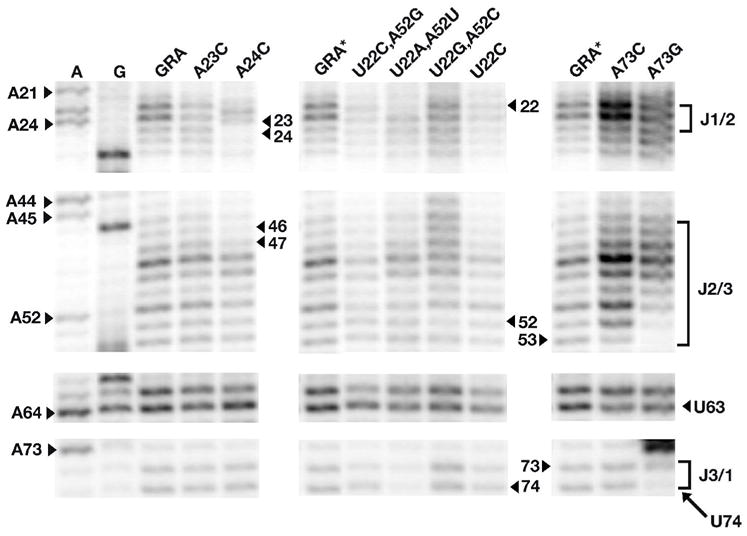
Probing the free state structure of three-way junction mutant GRA RNAs using N-methylisatoic anhydride (NMIA). Mutants are grouped according to their location in the three-way junction and shown in order of decreasing affinity for DAP (left to right). The lane corresponding to the wild type GRA shown at far left appears only once on the full sequencing gel (Supporting Figure 3) but is shown accompanying each set of mutants (GRA*) in order to illustrate the affect of each mutation; thus all three lanes labeled GRA are the identical lane from the gel. RNA was sequenced by inclusion of ddTTP or ddCTP in a reverse transcription reaction containing GRA to assign the nucleotide position of each modification (note that NMIA modification and the sequencing ladder are offset by one residue). Residues that are labeled correspond to the location of mutations and regions of possible alternative structure formation in the free state that prevents ligand binding in some mutants.
