Abstract
The field of molecular genetics has many roles in biodiversity assessment and conservation. I summarize three of those standard roles and propose logical extensions of each. First, many biologists suppose that a comprehensive picture of the Tree of Life will soon emerge from multilocus DNA sequence data interpreted in concert with fossils and other evidence. If nonreticulate trees are indeed valid metaphors for life's history, then a well dated global phylogeny will offer an opportunity to erect a universally standardized scheme of biological classification. If life's history proves to be somewhat reticulate, a web-like phylogenetic pattern should become evident and will offer opportunities to reevaluate the fundamental nature of evolutionary processes. Second, extensive networks of wildlife sanctuaries offer some hope for shepherding appreciable biodiversity through the ongoing extinction crisis, and molecular genetics can assist in park design by helping to identify key species, historically important biotic areas, and biodiversity hotspots. An opportunity centers on the concept of Pleistocene Parks that could protect “legacy biotas” in much the same way that traditional national parks preserve special geological features and historical landmarks honor legacy events in human affairs. Third, genetic perspectives have become an integral part of many focused conservation efforts by unveiling ecological, behavioral, or evolutionary phenomena relevant to population management. They also can open opportunities to educate the public about the many intellectual gifts and aesthetic marvels of the natural world.
Keywords: classification, nature reserves, phylogenetics
Creationis telluris est gloria Dei ex opere Naturae (The Earth's creation is the glory of God as seen in Nature's works).
Carolus Linnaeus, preface to Systema Naturae
In the 1700s, Carolus Linnaeus (1) devised a hierarchical system to rank and classify organisms. He did so without knowledge of evolution, presuming instead that static species (albeit modified occasionally by hybridization) had been present since the time of Creation. A century later, Charles Darwin (2) identified natural selection as a creative but natural agent of adaptive evolution. He did so without a proper understanding of genetics, sometimes presuming that heredity involved miscible gemmules in the blood. Nearly a century later, in the mid-1900s, Aldo Leopold (3) crafted a powerful environmental ethic based on ecological considerations. The extraordinary accomplishments of these three great scientists illustrate that systematics, evolutionary biology, and conservation science—three cornerstones of modern biodiversity research—can be (and often have been) practiced successfully without material input from the field of genetics. This is ironic because, fundamentally, evolution is genetic alteration through time, biodiversity is genetic diversity (including epigenetic and emergent phenomena), and nature's genetic diversity is what is being depleted in the current extinction crisis that has spurred the conservation movement.
A growing awareness of genetic operations and principles, beginning with the findings of Gregor Mendel (a younger contemporary of Darwin), contributed hugely to the “modern evolutionary synthesis” in the mid-1900s (4–6). Nevertheless, until the 1960s at least, organismal phenotypes (such as various morphological and behavioral traits) continued to provide the vast majority of empirical data for biodiversity research. Only in the last half-century have biologists gained extensive direct access to the hereditary information embedded in the molecular structures of nucleic acids and proteins (7, 8). What have these molecular genetic data added to the evolutionary synthesis and to conservation efforts?
Much of the molecular revolution in evolutionary biology has focused on mechanistic connections between genotype and phenotype, i.e., on attempts to understand “the genetic basis of evolutionary change” (9). In particular, a relatively young but burgeoning field known as evolution-development (“evo-devo”) addresses how the evolving genomes of diverse taxa are epigenetically modified and otherwise regulated during ontogeny to yield particular organismal phenotypes, including complex adaptations (10, 11). The evo-devo paradigm will continue to motivate scientific interest and generate vast research opportunities for the foreseeable future.
Here, I discuss three other areas of opportunity for molecular genetics in evolutionary biology, specifically in the realms of phylogenetics and conservation. For each of these three topics in a discipline that I call “biodiversity genetics,” I first summarize conventional wisdom, but then I intend to be provocative by raising scientific proposals that currently are far from mainstream but nevertheless have the potential to invigorate and perhaps even reshape the biodiversity sciences.
Tree of Life
Background.
Legions of biologists are currently gathering extensive molecular genetic data as part of a grand collective effort to reconstruct, once and for all, the history of life on Earth (12). Guiding this endeavor is the powerful conceptual metaphor of a phylogenetic tree, popularized by Ernst Haeckel in 1866 (13). Within the next decade or two, major branches and numerous twigs in the Tree of Life will be reconstructed (nearly as accurately as may ever become possible given the finite size of genomes and the relative ease by which DNA sequence data can now be gathered). The Tree of Life project will have completed its initial descriptive mission when it enters a more mature phase in which the gains in phylogenetic understanding about species' relationships, per unit of sequencing effort, will gradually diminish.
In the meantime, goals of the exuberant young Tree of Life initiative are to estimate not only branch topologies but also the evolutionary dates of various internal nodes (14). Molecular “clocks” will play a key role. Rates of DNA sequence evolution are known to be highly variable across lineages and loci (15), but experience indicates that when clocks are carefully calibrated and the dates they imply are compiled across dozens or even hundreds of unlinked loci, approximate origination times can be recovered for particular clades (16, 17). The calibrations normally require secure temporal reference points from independent evidence, e.g., from paleontology or biogeography. Thus, estimating absolute dates as well as branch topologies in phylogenetic trees is inherently an integrative endeavor that should engage many of the biodiversity sciences.
The Tree of Life project per se will be much like the Human Genome project, merely the first step in a vastly broader research agenda. The complete nucleotide sequence of a human genome provided a foundation for investigating genomic operations more deeply, such as enabling geneticists to map and characterize the structures and functions of genes responsible for particular phenotypes. Analogously, a robust Tree of Life will provide a foundation for delving much deeper into nature's evolutionary operations, such as enabling phylogeneticists to map the origins and evolutionary transitions among particular organismal phenotypes or document instances of reticulate evolution (18). All of these sentiments are simply conventional wisdom.
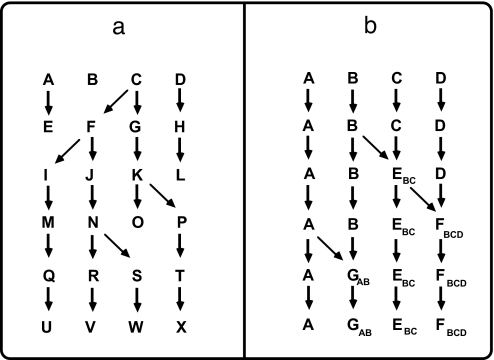
I suggest that either of two polar-opposite patterns (or more likely some mixture of the two) could emerge from the Tree of Life project and that either outcome would offer its own unprecedented grand opportunity for the field of evolutionary biology. One possibility is that the tree metaphor will apply well to many or most taxonomic groups, in which case an opportunity (described later) would arise for the field of systematics. Alternatively, the tree metaphor may prove to be inadequate, and an anastomose web or network of life would better describe the histories of descent of many taxa. Several recent authors have argued that genetic exchanges across lineages, via endosymbiotic mergers and lateral DNA transfers especially in microbes (19–21) or via hybridization in metazoan plants and animals (22), have played important evolutionary roles. For example, McCarthy (23) builds a case that new species seldom arise from the standard population genetic processes of gradual divergence via mutation, drift, and selection in allopatry, but instead that novel life forms often originate via the genetic stabilization of recombinant lineages following hybridization events (Fig. 1). If this hypothesis is correct, the ramifications for many areas of evolutionary biology would be profound (as described later).
Fig. 1.
Simplified depictions of two competing hypotheses about the genetic history of life. (a) The tree model characteristic of traditional phylogenetic thought. (b) An example of a network model [as advanced by McCarthy (23) in the context of hybridization and stabilization of recombinant genotypes]. Uppercase letters indicate different species or phenotypically recognizable life forms, arrows indicate historical pathways of descent, and successively lower rows in the diagrams represent more recent horizons in evolutionary time. When viewed backward in time, lineages shrink or coalesce to particular ancestors in a tree, but they expand to multiple ancestors in a network (because each new species is of hybrid origin).
Deciding whether the tree metaphor or the network metaphor betters explains the history of life is a stiff challenge requiring detailed and critical appraisals of empirical evidence for many taxonomic groups. But the two hypotheses do have several distinct predictions. In terms of genealogical expectations, for example, the Tree of Life model predicts that gene trees should be topologically concordant with one another and with the species tree they compose [barring potential complications such as insufficient resolution, hemiplasy (idiosyncratic lineage sorting across successive nodes in a species phylogeny) (24), and homoplasy]. In contrast, the network of life model predicts that multiple gene trees in a given taxonomic group will often be qualitatively discordant with one another (even after factoring out the complications of homoplasy and hemiplasy) because different DNA sequences may genuinely have highly distinct organismal histories. In evaluating the evidence, biologists must keep an initial open mind with regard to the network model because, under the competing tree model, a subtle danger exists of circular logic: Any comparative dataset can be used to reconstruct a phylogenetic tree when a tree provides the suppositional metaphor for the data analysis. Even inanimate entities (such as different kinds of chairs or cars) can be grouped into tree-like depictions based on their similarities or differences.
Depending on which evolutionary metaphor (branched genealogical trees or anastomose networks) proves generally correct for life's history, important but entirely different assignments will then emerge for the next generation of evolutionary biologists.
Assignment Given the Tree Model: Develop a Standardized Classification Scheme.
The two basic functions of biological taxonomy are to (i) provide a universal system for information storage and retrieval, and (ii) encapsulate an evolutionary interpretation of biological diversity (25). Unfortunately, current biological classifications are grossly nonstandardized because: (i) the species in named taxa are typically united by some unspecified mix of similarity by resemblance and similarity by descent, and (ii) even when the nested taxonomic ranks in a Linnaean hierarchy do register bona-fide nested clades the rankings remain noncomparable across different kinds of organisms (because no serious attempt has ever been made to normalize assayed characters, equilibrate taxonomic assignments, or even adopt any standardized criteria for taxonomic ranking). For example, some taxonomic genera such as Drosophila are an order of magnitude older than others such as Gorilla or Pan, and, because of an apples-versus-oranges problem, a taxonomic rank (such as a genus) shared by fruit flies and primates implies nothing about whether such taxa are similar with respect to genetic, phenotypic, or any other aspect of evolutionary diversity. As noted by De Queiroz and Gauthier (26), “No scientific enterprise, least of all one that considers the promotion of nomenclatural universality as one of its primary objectives, can accept the inconsistencies and ambiguities current in biological taxonomy.” Or as phrased by Hennig (27), “If systematics is to be a science it must bow to the self-evident requirement that objects to which the same label is given must be comparable in some way.”
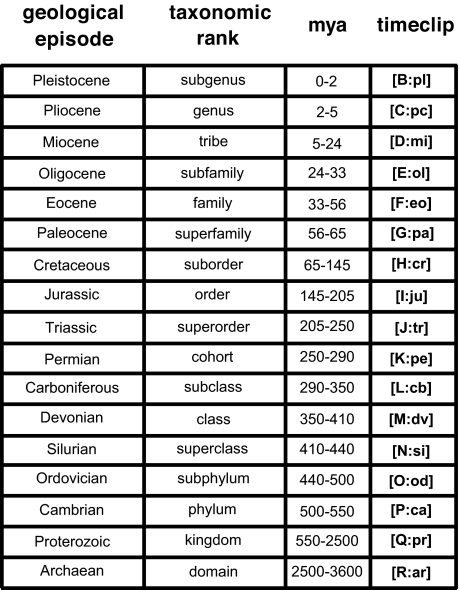
This state of affairs could, in principle, be rectified if systematists were to adopt absolute geological time as the universal evolutionary yardstick against which to standardize taxonomic assignments for extant clades of known age. The basic idea, proposed by Hennig (27) and elaborated by Avise and Johns (28), is that extant species that separated from a common ancestor in a specified window of evolutionary time would be assigned a taxonomic rank defined by that temporal band. The boundaries of the temporal windows are arbitrary at the outset and must be ratified by convention, but a proposal that I favor in principle would link each taxonomic rank to a specific geological episode. Serendipitously, there are 17 supraspecific ranks in modern versions of the Linnaean hierarchy (29) and also 17 primary subdivisions in the traditional geological time scale (30), thus affording the possibility of a perfect one-to-one allocation of taxonomic rank to geological episode (Fig. 2).
Fig. 2.
Examples of how a strategy of temporal banding might be used to standardize biological classifications for extant species (see text). Shown is the one-to-one correspondence possible between 17 standard taxonomic ranks in some modern versions of the Linnaean hierarchy (see ref. 29) and the temporal bands (Mya) for 17 traditionally recognized geological episodes (see ref. 30). In one temporal-banding proposal, current classifications and nomenclatures could be revised (perhaps drastically), such that each clade would be ranked and named strictly according to the temporal window in which it arose. Under a less drastic proposal (which I favor), current classifications and nomenclatures would be retained, but each existing taxonomic name would simply be appended with a time clip signifying the approximate date of that taxon's origination. Note that these temporal-banding proposals do not extend to species-level taxonomic assignments, where biological criteria, including reproductive isolation (regardless of a species' date of evolutionary origin), would continue to apply.
If the field of systematics from its outset had been able to implement a temporal-banding strategy for erecting biological classifications, many of the inconsistencies and ambiguities in current taxonomies could have been avoided. But formal biological names and classifications have their own historical legacies that cannot be ignored, and taxonomic stability also is highly important in systematics. One way to circumvent name changes and yet still implement the temporal-banding philosophy would be to attach a time clip (Fig. 2) to each extant taxon for which a reliable date of origin has been established from molecular-genetic or other evidence (31). For example, the familiar generic names Drosophila and Pan could be retained and merely time-clipped (with F:eo and C:pc) to signify their highly different evolutionary ages. Extant taxa for which origination dates remain unknown would lack time clips, but this too would convey important information by notifying the reader that a taxon's evolutionary age might be a worthy topic for additional investigation. After time clips become available for many organismal groups, it would be a simple matter for anyone to identify, sort, and compare even disparate kinds of taxa according to their approximate dates of evolutionary origin.
A temporal-banding scheme (especially as implemented in the time-clip format) could offer systematics and the biodiversity sciences several substantial benefits (elaborated in refs. 28 and 31). It would standardize biological classifications and thereby dramatically increase their comparative information content. It would both foster and facilitate comparisons of evolutionary rates in numerous genetic and phenotypic attributes (because absolute time is the denominator in any rate equation, and the time-clipped taxon names would specify approximate dates of clade origin). It would retain the well established Linnaean ranking system, including familiar taxonomic names, yet simultaneously enable systematists to incorporate substantive new phylogenetic knowledge, as it becomes available, into a biological classification. It would promote the often neglected notion that every phylogenetic tree has a temporal as well as a cladistic dimension and that both are important subjects for investigation. It should engage and foster collaborations among many of the biodiversity sciences in a community-wide phylogenetic mission to chart and interpret the temporal as well as cladogenetic dimensions of the planet's evolutionary heritage.
Assignment Given the Network Model: Reconsider the Nature of Evolutionary Processes.
If the network model (e.g., Fig. 1b) proves to be more nearly correct for many taxonomic groups, then the challenges for systematics and evolutionary biology will be entirely different (23). First, phylogeneticists would have to admit that their dream of reconstructing a branched tree of life had been merely a pipedream, and they would have to accept the new and probably far more difficult challenge of working out the precise history of reticulation events for each organismal group and how such reticulate genealogical histories have idiosyncratically distributed particular bits and pieces of DNA from disparate sources to extant taxa. Traditional concepts of species, phylogeny, ancestry, and classification, as well as the significance of reproductive isolation, would all have to be reevaluated. Biologists would have to embrace the notion that biological processes falling somewhat outside the standard neo-Darwinian paradigm for speciation (such as interspecific hybridization and the reproductive stabilization of genetic-recombinant derivatives) could play major and previously underappreciated roles in evolution. They would have to reevaluate the origins of genetic variation on which natural selection acts and how novel phenotypic adaptations and different forms of life mechanistically come into being. In short, major shifts in evolutionary thought would be required, and this would open wonderful opportunities for the eventual emergence of a grandly updated evolutionary synthesis, 21st-century style.
Pleistocene Parks
… suppose that the United States and the other leading developed countries could agree on a regular allocation for global biodiversity protection so that billions of dollars, rather than millions, could annually flow into parks and park protection. What then?
John Terborgh (32)
Background.
In an eloquent requiem for nature, Terborgh (32) has argued that, in the face of a globally burgeoning human population, the only credible prospect for preserving substantial biodiversity will be for governments [or other entities such as nongovernmental organizations (NGOs)] to set aside extensive nature sanctuaries and then actively protect those parklands in perpetuity. Many countries, including the United States, have long established systems of National Parks that usually feature special landscapes and geological formations (such as the picturesque rocky shores of Acadia Park in Maine, the majestic mountains of Glacier Park in Montana, or the special volcanic features of Yellowstone Park in Wyoming). A growing realization is that analogous and extensive reserve systems across the globe also are urgently needed to offer explicit protection for the biological world's special features, such as endangered species, distinctive biotic communities and ecosystems, and biodiversity “hotspots” (33, 34).
Accordingly, many scientists and conservation organizations are actively engaged in identifying threatened sites around the world where exceptional concentrations of rare or endemic species still exist and where conservation efforts might therefore be focused to best effect (35–37). For example, it has been estimated that as many as 44% of vascular plant species and 35% of all vertebrate species (exclusive of fishes) are confined to 25 biodiversity hotspots that comprise only 1.4% of Earth's land surface, and that for the cost of perhaps as little as $500 million annually, a biotic reserve system centered on such treasure-rich locations could be a “silver bullet” for biodiversity protection (35). A related suggestion is that sites meriting high priority for protection should display exceptional concentrations of phylogenetically distinctive taxa (38–42), the rationale being that organismal lineages with long-independent evolutionary histories contain disproportionately large fractions of the planet's total extant genomic biodiversity (43–45).
These various suggestions for biotic reserves need not be at odds. Indeed, given the dire prospects for global biodiversity in the ongoing extinction crisis and the total inadequacy to date of commensurate responses by most governments, the more natural parklands that societies can be persuaded to sequester under any reasonable biological motivation, the better. Furthermore, the parkland effort need not be confined to governmental initiatives, as well illustrated by the welcome activities of NGOs such as the Nature Conservancy and Conservation International. A related hope is that philanthropists and profit industries also will become increasingly persuaded of the urgency to protect remaining nature, if for no other reason than in their own enlightened financial (as well as ethical) self-interest.
To pick just one such example of the potential for private involvement, an inspirational business venture (“IQ RESORTS by PANGAEA WORLD”) spearheaded by Hana Ayala (46) aims to partner responsible and forward-thinking members of the hotel/tourism industry with world-class scientists in a global vision to promote science and protect biodiversity as an integral part of the business plan (which would include the acquisition and preservation of extensive nature reserves in key locations, as well as the generation of new funding mechanisms for the biodiversity sciences). Three underlying premises of this initiative are as follows: (i) knowledge mobilized through scientific research is the ultimate inexhaustible resource; (ii) the world's most spectacular and biodiverse landscapes and seascapes are primary reservoirs for scientific knowledge that in turn can promote long-term conservation efforts in pragmatically effective and economically sustainable ways; and (iii) the international hotel industry—with its collective global ambition and growing emphasis on mind-stimulating travel experiences—has perhaps more financial interest, capacity, and incentive than any other private industry to partner with science in charting and protecting the world's premier biological heritage reserves. The PANGAEA initiative aims to go well beyond traditional nature-tourism ventures by envisioning a global archipelago of interconnected “wonder sites” where the scientific study and preservation of nature are the explicit and formal motivation for linking sustainable economics with science.
The perspectives and data of ecological and evolutionary genetics can contribute to parkland conservation efforts in many ways. For example, they can help to identify species and biodiversity hotspots, especially for otherwise poorly known taxonomic groups. They can vastly improve our understanding of phylogenetic relationships of numerous taxa within and among the extant regional biotas that conservationists might seek to protect (7, 47, 48). Finally, they can help to illuminate many management-relevant aspects of the biology and natural history of particular species that warrant special conservation concern.
Most of the general sentiments summarized above (a notable exception perhaps being the PANGAEA WORLD initiative) reflect conventional wisdoms, at least among many biologists. Here I suggest how phylogeographic perspectives might offer an additional opportunity in parkland motivation that is less widely appreciated. Phylogeography is a relatively young biological field that deals with descriptions and interpretations of the spatial distributions of genealogical lineages, especially within and among closely related species (49). An emerging phylogeographic generality is that many, if not most, extant taxonomic species are spatially subdivided into small numbers of highly distinctive historical units (50).
Many of these distinctive genealogical entities [sometimes referred to as evolutionarily significant units (ESUs)] (51, 52) began diverging from one another in unglaciated biological refugia of the Pleistocene Epoch or earlier (53–55). In Europe, for example, extant populations of many plant and animal species bear the genomic footprints of phylogeographic differentiation in several disjunct ice-free areas (notably the Iberian Peninsula, the Italian Peninsula, and the Balkans) typically followed by post-Pleistocene dispersal from one or more of these ancestral homelands (56–58). Likewise, key genealogical separations presumably tracing back to historical refugia distinguish regional populations of many species in different sections of the eastern United States (59, 60). Qualitatively similar patterns also have been uncovered in comparative phylogeographic surveys of regional biotas in several other parts of the world (49, 61). In at least several cases, the current boundaries between ESUs tend to be spatially concordant with transition zones between zoogeographic provinces as identified by more traditional evidence (such as species' ranges and faunal distributions). Such concordance suggests that similar types of evolutionary forces (perhaps operating as detailed in ref. 49) may be responsible for both of these seemingly unrelated biogeographic phenomena.
Assignment: Identify and Preserve Nature's Recent Historical Legacies.
The phylogeographic observations discussed above suggest that a concept—of Pleistocene Parks or Phylogeographic Sanctuaries—might be added to the compelling list of scientific rationales for earmarking particular regional nature reserves. Such nature reserves (like those based on traditional biodiversity hotspots) would protect and highlight the distinctive “legacy biotas” they contain, in much the same way that traditional historical landmarks (such as Civil War battlegrounds) honor important legacy events in human affairs. A carefully designed archipelago or network of phylogeographic reserves on each continent and in each marine region could thus add an emotive element of historical legacy to the catalog of societal inducements to preserve biodiversity. Furthermore, a widely promoted concept of Pleistocene Parks (like the evocative notion of Jurassic Park) might resonate well with the public and policymakers. It also might dovetail nicely with the PANGAEA WORLD initiative discussed above and perhaps also with proposals to “re-wild” ecosystems with Pleistocene-like biotas (62).
Thus, a compelling assignment for the field of comparative phylogeography will be to map the spatial and temporal dimensions of Earth's remaining genealogical capital on all of the world's continents and ocean regions (a task already well initiated in several areas, such as Europe and parts of North America). A comprehensive phylogeographic inventory of Earth's microevolutionary history will complement ongoing attempts to identify and catalog all extant species (see ref. 63), and it also will complement ongoing appraisals of Earth's macroevolutionary history in the Tree of Life project. An overarching practical mission will be to incorporate information from all of these integrative endeavors into meaningful conservation plans, notably with regard to implementing the concept of regional sanctuaries for nature (64).
Biodiversity Education
Ultimately, nature and biodiversity must be conserved for their own sakes, not because they have present utilitarian value … the fundamental arguments for conserving nature must be spiritual and aesthetic, motivated by feelings that well up from our deepest beings.
John Terborgh (32)
Background.
“Conservation genetics” has become a popular discipline, as evidenced, for example, by two edited compilations (65, 66), a teaching textbook (67), and a scientific journal (initiated in 2001), all bearing within their titles that exact two-word phrase. Historically, the field was associated mostly with studies of inbreeding depression and the loss of heterozygosity in small populations, but its purview has expanded greatly in recent years to include a wide range of empirical and theoretical studies that basically attempt to illuminate how patterns of genetic diversity are distributed within and among individuals, kinship groups, populations, species, and supraspecific taxa (68). Such investigations (typically using molecular markers) routinely include genetic appraisals of the following: plant and animal mating systems, behaviors, and natural histories; magnitudes and patterns of population structure due to past and present demographic factors; gene flow, genetic drift, and various categories of natural selection; other evolutionary phenomena such as patterns and processes of speciation, hybridization, introgression, and phylogenetics; forensic analyses of wildlife and wildlife products; and many additional genetic topics that are often highly germane to the principles and the practice of conservation biology.
All of these sentiments are standard wisdom among modern biologists. So too is the realization that a strong societal preference exists for saving species that are large, attractive, or emotionally evocative, compared with those that are small, drab, or unobtrusive. Almost inevitably, conservation efforts thus become biased toward “charismatic megabiota” (69). I suggest another role for conservation genetics that is somewhat more amorphous, but nevertheless has a huge potential to elicit additional public support for meaningful societal action on behalf of nature and biodiversity protection. I am referring to a compelling educational mission: to enthuse students of all ages, including the general public as well as political, social, and religious leaders, about nature's countless underappreciated marvels.
Nearly all creatures (including the “charismatically challenged”) have fascinating natural-history stories to tell, and scientists as well as natural theologians for centuries have delved into nature's workings through field observations and phenotypic investigations. Yet before the advent and widespread use of molecular markers, many of nature's incredible operations remained hidden from view. Nature can now be revealed at and through this new window of molecular-level observation, and the results are often far more engrossing than might ever have been predicted.
First are the astounding findings about genomes. Even a few years ago, few scientists could have imagined that genes encoding functional RNA and protein molecules of obvious benefit to the organism would prove to constitute only a small fraction of the eukaryotic genome, and that the rest of the composite DNA sequence includes an astonishing collection of noncoding regions, regulatory modules, pseudogenes, and legions of repetitive elements, many of which are descended from selfish virus-like elements that have proliferated and jumped around the genome often at the immediate fitness expense of their hosts. A new metaphor is emerging in which each eukaryotic genome can be viewed, in effect, as a miniature ecological community whose quasi-independent members (unlinked DNA sequences) all struggle for representation in the next generation of sexual reproducers and thereby become involved in elaborate coevolutionary games that can be quite analogous to the parasitisms, commensalisms, and mutualisms routinely observed among species in natural ecosystems (70). This metaphor of the genome as a submicroscopic community of genes constantly undergoing evolutionary adjustments is far from perfect, but it does promote a perspective on genomic operations that today may be much more useful and research-stimulating than earlier genomic metaphors (such as the “beads on a string” image of functional and fully collaborative genes packed tightly along chromosomes).
A second arena in which molecular genetic markers are having a huge scientific impact is in uncovering heretofore hidden secrets about the ecologies, behaviors, natural histories, and evolution of organisms in nature. An adequate discussion of this topic is far beyond the scope of this article, so what follow are merely a few examples of the many types of questions that scientists have answered using molecular markers, but that for one logistical reason or another had been inadequately addressed by earlier field observations or phenotypic assessments. For fuller answers to the following questions and many others like them, all in layperson language, see refs. 18 and 71.
How big and old can natural clones of mushrooms become? (Living members of one clone were estimated to weigh a collective 100 tons, occupy 40 acres, and derive from a single zygote that formed ≈1,500 years ago.) Does each female green turtle (a highly migratory marine species) return to her natal beach to nest? (Yes, normally.) Why do female roly-poly pill bugs often greatly outnumber males? (Because many strains are infected by intracellular parasitic bacteria that are maternally transmitted and, accordingly, have evolved the physiological capability to transform male roly-polys into functional females.) Does a pregnant male pipefish or seahorse often carry a brood of offspring from more than one dam? (In some species, yes; in other cases, no.) What fraction of embryos in the nests of bluegill sunfish are foster progeny attributable to cuckoldry by sneaker males? (Approximately 20% in one well studied population.) Did the bipedal hop arise once or multiple times in kangaroos evolution? (Probably once only, according to phylogenetic analysis.) Why do king crabs have an asymmetrically twisted abdomen? (Because this trait appears to be a phylogenetic legacy retained from hermit crab ancestors whose coiled abdomens had evolved to fit nicely into deserted snail shells that hermits adopt as protective homes.) Which came first in evolution, the chicken or the egg? (The hard-shelled egg came first, by ≈300 million years.)
Assignment: Educate the Public to Nature's Marvels.
In the end, we conserve only what we love. We will love only what we understand. We will understand only what we are taught.
Baba Dioum, Senegalese poet.
A sad predicament for conservation efforts in the modern world is that a large fraction of humanity is estranged from nature, a situation that is likely to get worse as urbanization increases and human numbers soar. For example, I teach at a major university most of whose undergraduate students come from the metropolitan Los Angeles basin, and relatively few of those students seem to have had much opportunity for substantive personal contact with nature. Furthermore, our biology curriculum offers few “organismal” courses that might help to alleviate this problem. The situation here in Southern California is hardly unique. How can educators enthuse their students about biodiversity when direct experiences with nature have not been a significant part of those students' upbringing?
The good news is that many students (as well as many members of the general public) seem willing and eager to embrace nature if simply given the opportunity. Therein lays a third grand mission for molecular genetics and the other biodiversity sciences in conservation efforts: to cultivate in students of all ages a sense of awe, respect, and appreciation for the numerous other creatures—including the charismatically challenged—that share our crowded and imperiled planet. As phrased by E. O. Wilson (72), “… to the degree that we come to understand other organisms, we will place a greater value on them, and on ourselves.” And, as noted by the late Stephen J. Gould (73), “We cannot win this battle to save species and environments without forging an emotional bond between ourselves and nature … for we will not fight to save what we do not love.”
An emotional and intellectual appreciation of nature, and also of rational scientific efforts to comprehend its workings, can be stimulated in many ways. Visual presentations (such as the Life on Earth TV series or the March of the Penguins movie) can play huge roles in educating the public. So too can eloquent thoughts and words, spoken or written. Fortunately, many biologists take delight in conveying the excitement of natural history and the joy of scientific inquiry to their students and also to the general public via trade books, lectures, service in conservation organizations, and other venues. Such efforts should be encouraged, applauded, and rewarded because only an educated public is motivated to demand a place for nature on this human-dominated planet.
Conclusion
The next few decades offer our best and last remaining chance to shepherd appreciable biodiversity through the current global extinction crisis. This monumentally important task should be at the forefront of societal consciousness and action not only because nature offers vast economic and material benefits to humanity, enriches our lives both aesthetically and intellectually, and provides bountiful scientific opportunities to understand the biological context of our existence. More basically, we should cherish nature because it is the ethically proper thing to do. Protecting what remains of nature must become our collective moral imperative. If it does not, we will lose not just nature herself, but also a deeply basic element of our humanity. We must come to value nature for nature's sake (as well as our own), instill that fundamental ethos in our children, and bequeath to future generations a planet that is no less biodiverse than the one into which we were born.
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution II: Biodiversity and Extinction,” held December 6–8, 2007, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS web site at www.nasonline.org/Sackler_biodiversity.
The author declares no conflict of interest.
References
- 1.Linnaeus C. Systema Naturae. New York: Wheldon and Wesley; 1759. reprinted (1964) [Google Scholar]
- 2.Darwin C. On the Origin of Species by Means of Natural Selection. London: John Murray; 1859. [Google Scholar]
- 3.Leopold A. A Sand County Almanac. New York: Oxford Univ Press; 1949. [Google Scholar]
- 4.Dobzhansky T. Genetics and the Origin of Species. New York: Columbia Univ Press; 1937. [Google Scholar]
- 5.Mayr E. Systematics and the Origin of Species. New York: Columbia Univ Press; 1942. [Google Scholar]
- 6.Stebbins GL. Variation and Evolution in Plants. New York: Columbia Univ Press; 1950. [Google Scholar]
- 7.Avise JC. Molecular Markers, Natural History, and Evolution. 2nd Ed. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 8.Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. 2nd Ed. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 9.Lewontin RC. The Genetic Basis of Evolutionary Change. New York: Columbia Univ Press; 1974. [Google Scholar]
- 10.Avise JC, Ayala FJ, editors. In the Light of Evolution: I. Adaptation and Complex Design. Washington, DC: Natl Acad Press; 2007. [PubMed] [Google Scholar]
- 11.Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. 2nd Ed. Malden, MA: Blackwell; 2004. [Google Scholar]
- 12.Cracraft J, Donoghue MJ, editors. Assembling the Tree of Life. Oxford: Oxford Univ Press; 2004. [Google Scholar]
- 13.Haeckel E. Generelle Morphologie der Organismen. Berlin: Georg Reimer; 1866. [Google Scholar]
- 14.Hedges B, Kumar S, editors. Timetrees of Life. Oxford: Oxford Univ Press; 2008. in press. [Google Scholar]
- 15.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 16.Kumar S. Molecular clocks: Four decades of evolution. Nat Rev Genet. 2005;6:654–662. doi: 10.1038/nrg1659. [DOI] [PubMed] [Google Scholar]
- 17.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 18.Avise JC. Evolutionary Pathways in Nature: A Phylogenetic Approach. Cambridge, UK: Cambridge Univ Press; 2006. [Google Scholar]
- 19.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 20.Margulis L, Sagan D. Acquiring Genomes: A Theory of the Origin of Species. New York: Basic Books; 2002. [Google Scholar]
- 21.Arnold MJ. Evolution Through Genetic Exchange. New York: Oxford Univ Press; 2006. [Google Scholar]
- 22.Arnold MJ. Natural Hybridization and Evolution. New York: Oxford Univ Press; 1997. [Google Scholar]
- 23.McCarthy EM. On the Origin of New Life Forms. New York: Oxford Univ Press; 2008. [Google Scholar]
- 24.Avise JC, Robinson TJ. Hemiplasy: A new term in the lexicon of phylogenetics. Syst Biol. 2008 doi: 10.1080/10635150802164587. in press. [DOI] [PubMed] [Google Scholar]
- 25.Mayr E. The Growth of Biological Thought. Cambridge, MA: Belknap; 1982. [Google Scholar]
- 26.de Queiroz K, Gauthier J. Phylogenetic taxonomy. Annu Rev Ecol Syst. 1992;23:449–480. [Google Scholar]
- 27.Hennig W. Phylogenetic Systematics. Urbana, IL: Univ of Illinois Press; 1966. [Google Scholar]
- 28.Avise JC, Johns GC. Proposal for a standardized temporal scheme of biological classification for extant species. Proc Natl Acad Sci USA. 1999;96:7358–7363. doi: 10.1073/pnas.96.13.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayr E, Ashlock PA. Principles of Systematic Zoology. New York: McGraw–Hill; 1991. [Google Scholar]
- 30.Futuyma DJ. Evolutionary Biology. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 31.Avise JC, Mitchell D. Time to standardize taxonomies. Syst Biol. 2007;56:130–133. doi: 10.1080/10635150601145365. [DOI] [PubMed] [Google Scholar]
- 32.Terborgh J. Requiem for Nature. Washington, DC: Island; 1999. [Google Scholar]
- 33.Myers N. Threatened biotas: “Hotspots” in tropical forests. Environmentalist. 1988;10:243–256. doi: 10.1007/BF02240252. [DOI] [PubMed] [Google Scholar]
- 34.Reid WV. Biodiversity hotspots. Trends Ecol Evol. 1998;13:275–280. doi: 10.1016/s0169-5347(98)01363-9. [DOI] [PubMed] [Google Scholar]
- 35.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 36.Prendergast JR, Quinn RM, Lawton JH, Eversham BC, Gibbons DW. Rare species, the coincidence of diversity hotspots and conservation strategies. Nature. 1993;365:335–337. [Google Scholar]
- 37.Williams P, et al. A comparison of richness hotspots, rarity hotspots, and complementary areas for conserving diversity of British birds. Conserv Biol. 1996;10:155–174. [Google Scholar]
- 38.Vane-Wright RI, Humphries CJ, Williams PH. What to protect: Systematics and the agony of choice. Biol Conserv. 1991;55:235–254. [Google Scholar]
- 39.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 40.Crozier RH. Preserving the information content of species: Genetic diversity, phylogeny, and conservation worth. Annu Rev Ecol Syst. 1997;28:243–268. [Google Scholar]
- 41.Krajewski C. Phylogenetic measures of biodiversity: A comparison and critique. Biol Conserv. 1994;69:33–39. [Google Scholar]
- 42.Humphries CJ, Williams PH, Vane-Wright RI. Measuring biodiversity for conservation. Annu Rev Ecol Syst. 1995;26:93–111. [Google Scholar]
- 43.May RM. Taxonomy as destiny. Nature. 1990;347:129–130. [Google Scholar]
- 44.May RM. Conceptual aspects of the quantification of the extent of biological diversity. Phil Trans R Soc London Ser B. 1994;345:13–20. doi: 10.1098/rstb.1994.0082. [DOI] [PubMed] [Google Scholar]
- 45.Avise JC. Phylogenetic units and currencies above and below the species level. In: Purvis A, Gittleman JL, Brooks T, editors. Phylogeny and Conservation. Cambridge, UK: Cambridge Univ Press; 2005. pp. 76–100. [Google Scholar]
- 46.Lempinen EW, editor. Ayala's passion for knowledge shines at AAAS event. Science. 2006;312:542. [Google Scholar]
- 47.Godfray HCJ. Linnaeus in the information age. Nature. 2007;446:259–260. doi: 10.1038/446259a. [DOI] [PubMed] [Google Scholar]
- 48.Purvis A, Gittleman JL, Brooks T, editors. Phylogeny and Conservation. Cambridge, UK: Cambridge Univ Press; 2005. [Google Scholar]
- 49.Avise JC. Phylogeography: The History and Formation of Species. Cambridge, MA: Harvard Univ Press; 2000. [Google Scholar]
- 50.Avise JC, Walker DW. Species realities and numbers in sexual vertebrates: Perspectives from an asexually transmitted genome. Proc Natl Acad Sci USA. 1999;96:992–995. doi: 10.1073/pnas.96.3.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryder OA. Species conservation and the dilemma of subspecies. Trends Ecol Evol. 1986;1:9–10. doi: 10.1016/0169-5347(86)90035-2. [DOI] [PubMed] [Google Scholar]
- 52.Moritz C. Comparative phylogeography and the identification of genetically divergent areas for conservation. Trends Ecol Evol. 1994;9:373–375. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 53.Avise JC, Walker DW. Pleistocene phylogeographic effects on avian populations and the speciation process. Proc R Soc London Ser B. 1998;265:457–463. doi: 10.1098/rspb.1998.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Avise JC, Walker DW, Johns GC. Speciation durations and Pleistocene effects on vertebrate phylogeography. Proc R Soc London Ser B. 1998;265:1707–1712. doi: 10.1098/rspb.1998.0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klicka J, Zink RM. The importance of recent Ice Ages in speciation: A failed paradigm. Science. 1997;277:1666–1669. [Google Scholar]
- 56.Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc. 1996;58:247–276. [Google Scholar]
- 57.Schmitt T. Molecular biogeography of Europe: Pleistocene cycles and postglacial trends. Front Zool. 2007 doi: 10.1186/1742-9994-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weiss S, Ferrand N, editors. Phylogeography of Southern European Refugia. Dordrecht, The Netherlands: Springer; 2007. [Google Scholar]
- 59.Avise JC. Molecular population structure and the biogeographic history of a regional fauna: A case history with lessons for conservation biology. Oikos. 1992;63:62–76. [Google Scholar]
- 60.Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS. Comparative phylogeography of unglaciated eastern North America. Mol Ecol. 2006;15:4261–4293. doi: 10.1111/j.1365-294X.2006.03061.x. [DOI] [PubMed] [Google Scholar]
- 61.Bermingham E, Moritz CC, editors. Comparative phylogeography. Mol Ecol. 1998;7:367–545. doi: 10.1046/j.1365-294x.1998.00358.x. (Special Issue) [DOI] [PubMed] [Google Scholar]
- 62.Martin PS. Twilight of the Mammoths: Ice Age Extinction and the Rewilding of America. Berkeley: Univ of California Press; 2005. [Google Scholar]
- 63.Blackmore S. Biodiversity update—progress in taxonomy. Science. 2002;298:365. doi: 10.1126/science.1075026. [DOI] [PubMed] [Google Scholar]
- 64.Moritz C, Faith DP. Comparative phylogeography and the identification of genetically divergent areas for conservation. Mol Ecol. 1998;7:419–429. [Google Scholar]
- 65.Loeschcke V, Tomiuk J, Jain SK, editors. Conservation Genetics. Basel, Switzerland: Birkhäuser; 1994. [Google Scholar]
- 66.Avise JC, Hamrick, editors. Conservation Genetics: Case Histories from Nature. New York: Chapman & Hall; 1996. [Google Scholar]
- 67.Frankham R, Ballou JD, Briscoe DA. Introduction to Conservation Genetics. Cambridge, UK: Cambridge Univ Press; 2002. [Google Scholar]
- 68.Avise JC. The history, purview, and future of conservation genetics. In: Carroll SP, Fox CW, editors. Conservation Biology: Evolution in Action. Oxford: Oxford Univ Press; 2008. in press. [Google Scholar]
- 69.Clark JA, May RM. Taxonomic bias in conservation research. Science. 2002;297:191–192. doi: 10.1126/science.297.5579.191b. [DOI] [PubMed] [Google Scholar]
- 70.Avise JC. Evolving genomic metaphors: A new look at the language of DNA. Science. 2001;294:86–87. doi: 10.1126/science.294.5540.86. [DOI] [PubMed] [Google Scholar]
- 71.Avise JC. Genetics in the Wild. Washington, DC: Smithsonian Inst Press; 2002. [Google Scholar]
- 72.Wilson EO. Biophilia. Cambridge, MA: Harvard Univ Press; 1984. [Google Scholar]
- 73.Gould SJ. Unenchanted evening. Nat Hist. 1991;100(5):7–14. [Google Scholar]