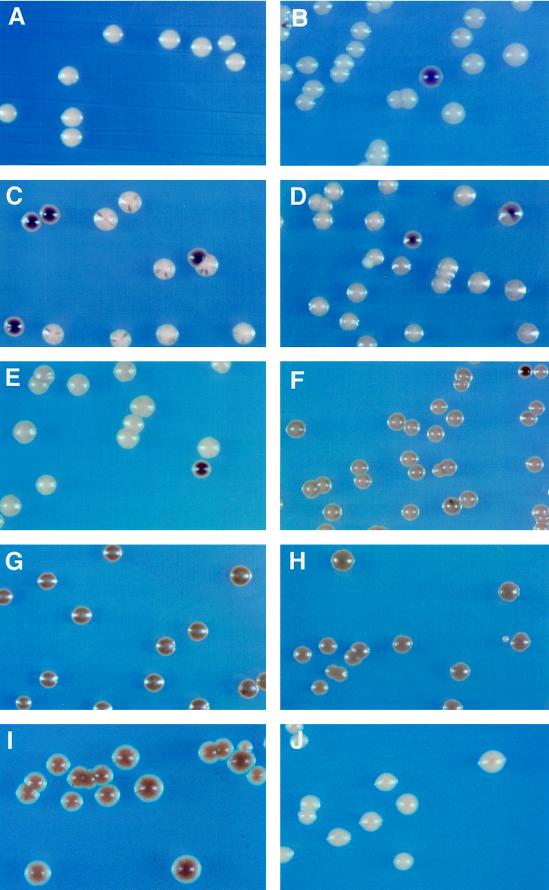
Figure 3.
Effect of various deletions on rDNA silencing. Cells from (A) M9 (Ty1-MET15 inserted in a non-rDNA locus), (B) M1 (RDN1::Ty1-MET15), (C) JS218 (M1 with sir2:: HIS3), (D) YSK757 (M1 with sir3Δ:: LEU2), (E) YSK753 (M1 with hda1Δ:: LEU2), (F) YSK781 (M1 with sir3Δ:: LEU2, hda1Δ:: HIS3), (G) YSK755 (M1 with rpd3Δ:: TRP1), (H) YSK783 (M1 with sir3Δ:: LEU2, rpd3Δ:: URA3), (I) YSK779 (M1 with rpd3Δ:: URA3, sir2:: HIS3), and (J) YCYL11 (M9 with rpd3Δ:: TRP1) were streaked on modified YPD agar medium containing Pb2+. The plates were incubated for 1 wk at 30°C. MET15+ cells grown on Pb2+ medium form white colonies (A), but met15 mutant cells develop dark brown colonies on the same medium. In the control M1 (B), the Ty1-MET15 is located upstream of the 5S rDNA in the RDN1 locus (Smith and Boeke, 1997).