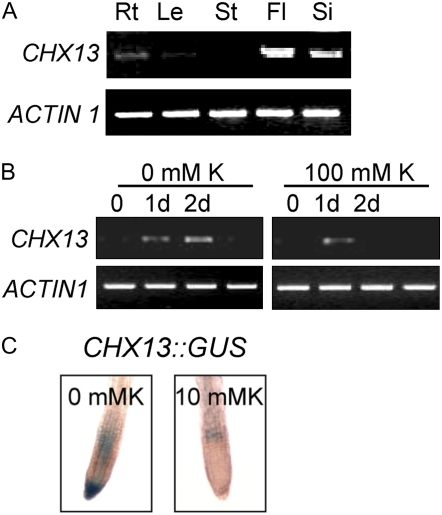
Figure 4.
AtCHX13 expression. A, Vegetative and reproductive organs. Total RNA was isolated from roots (Rt), leaves (Le), and stems (St; from 3-week-old plants), flowers (Fl), and siliques (Si; from 4-week-old plants), reverse transcribed, and subjected to RT-PCR. ACTIN1 was used as internal standard. Images shown represent results obtained from three independent amplification reactions. B, RT-PCR analysis of AtCHX13 expression when seedlings were grown on high and low K+. RT-PCR analysis of AtCHX13 expression. One-week-old Col-0 seedlings were transferred to 0.5× Murashige and Skoog medium supplemented with 100 mm KCl or to modified 0.5× Murashige and Skoog with low levels of K+ (the agar provides the medium with approximately 0.4 mm K+). The seedlings were collected at specified time points as indicated for RNA isolation. The samples were then analyzed as described in A. ACTIN1 was used as internal control. C, AtCHX13∷GUS lines were grown on 0.5× Murashige and Skoog plate for 9 d and then transferred to 0.5× Murashige and Skoog containing either no K+ or with 10 mm K for 2 d prior to GUS staining. [K+] was removed (0 mm K) by replacing KNO3, KH2PO4, and KI with BTP-NO3, NaH2PO4, and NaI, respectively.