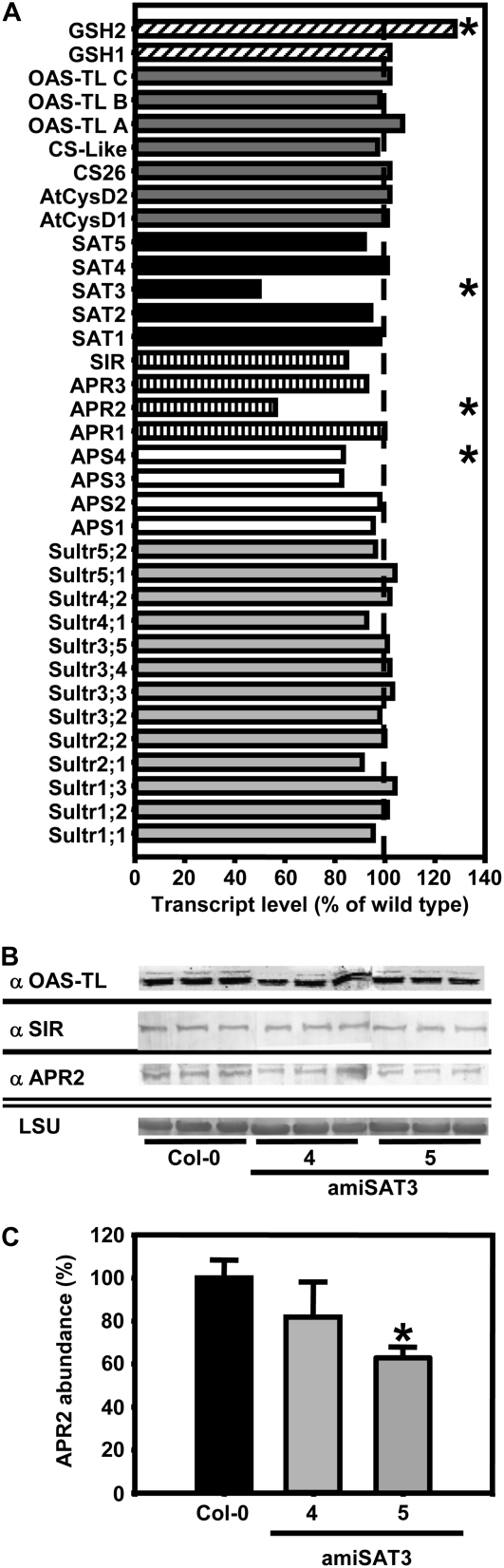
Figure 7.
Protein abundances and transcript levels of sulfur metabolism-related genes in leaves of transgenic Arabidopsis plants. A, The transcript levels of sulfur metabolism-related genes in leaves of amiSAT3 line 5 and wild-type Arabidopsis plants were compared using a targeted microarray approach. Total mRNA was extracted form three individuals of each plant line, labeled independently two times with Cy3 and Cy5, and cohybridized with the microarray two times (n = 11). The transcript levels of sulfate transporters (gray bars), ATP-sulfurylases (white bars), sulfate-reducing genes (dashed bars), SATs (black bars), OAS-TLs (dark gray bars), and genes participating in glutathione (GSH) synthesis (inclined dashed bars) in amiSAT3 line 5 are shown as percentages of wild-type levels. B, Proteins were extracted from three individuals of wild-type plants and amiSAT3 lines 4 and 5 of the same age. The abundances of OAS-TL A, B, and C, SIR, and APR2 were determined by immunoblotting using specific antibodies from crude extracts (15 μg). The large subunit of ribulose-1,5-bisphosphate carboxylase (LSU) served as a loading control after staining with Coomassie Brilliant Blue. C, The reduction of APR2 protein in amiSAT3 lines 4 and 5 (gray bars) in comparison with wild-type plants (black bars) was quantified densitometrically. Error bars indicate sd, and asterisks mark significant differences using the unpaired t test.