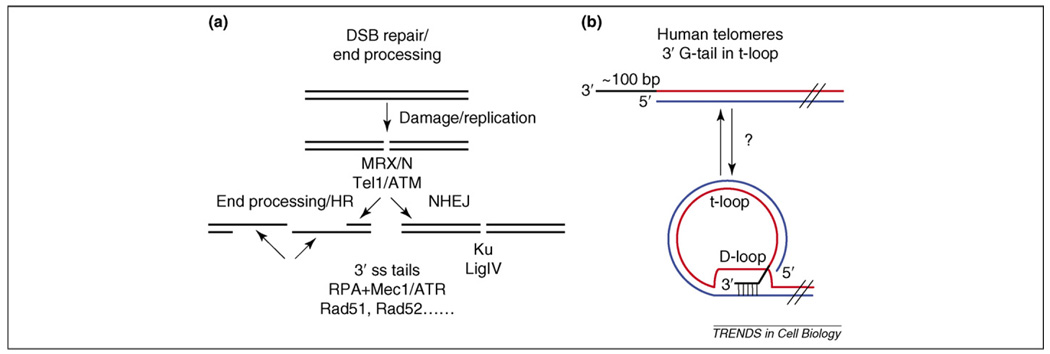
Figure 1.
Telomere structure and DSB processing. (a) A simplified version of the events that occur after the formation of a DNA double-strand break (DSB). The ends are bound by the MRX/N complex, which recruits the Ser–Thr protein kinase Tel1 (also known as ATM). Subsequent processing and protein binding will convert the break into either long stretches of single-stranded DNA that are a substrate for binding of RPA and recruitment of Mec1 (also known as ATR) before homologous recombination (HR) (left) or shuttle it down the Ku-dependent non-homologous end joining (NHEJ) pathway (right). (b) Mammalian telomeres have long G-tails (~75–300 bases) at both chromosome ends throughout the cell cycle – whether or not the cells express telomerase. By contrast, S. cerevisiae telomeres have short (~15 base) G-tails throughout most of the cell cycle, except in late S–G2 phase, when G-tails are much longer [16,101]. Long G-tails in S. cerevisiae are also telomerase-independent [16]. In mammals and other organisms (but probably not in S. cerevisiae), the G-tail can invade the duplex region of the telomere, displacing an internal segment of single-stranded G-telomeric DNA and forming a terminal circle. In T-loops, the G-tail is base-paired to the C-strand within an otherwise duplex region of telomeric DNA, displacing the G-strand in the original duplex and generating an internal stretch of ~100 bases of single-stranded G-strand telomeric DNA called a D-loop. T-loops are thought to protect the end against degradation and other hazards.