Figure 1.
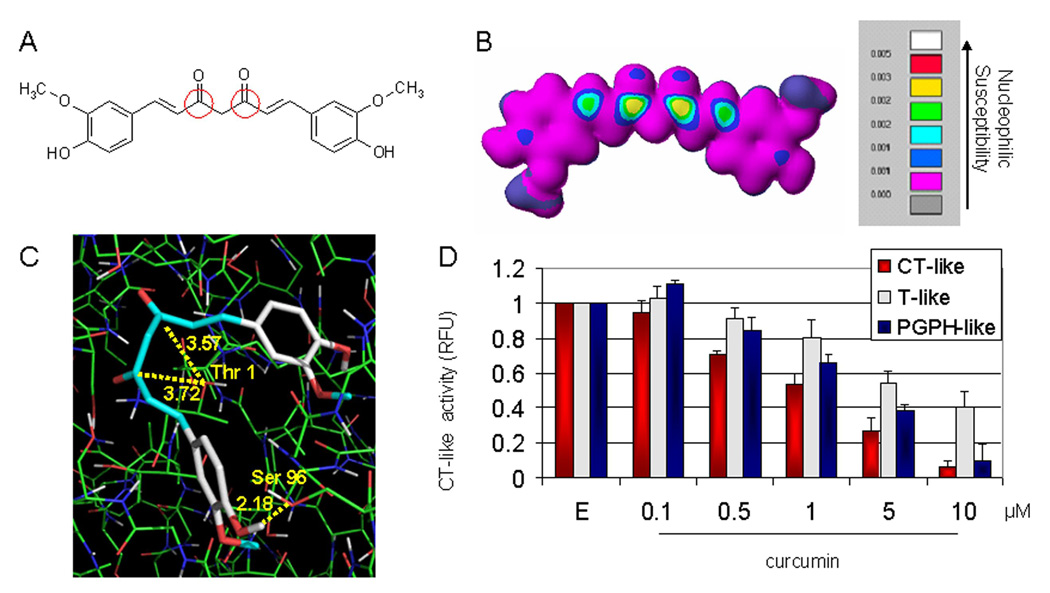
In silico and in vitro proteasome inhibition by curcumin. A, the chemical structure of curcumin. The regions with carbonyl carbons that have the SAR (structure activity relationship) are marked with red circles. B, molecular orbital energy analysis is demonstrated by drawing and electron density isosurface and coloring by nucleophilic susceptibility. The yellow center signifies the highest area of susceptibility. C, docking analysis of curcumin, which is represented by the stick structure and the colors are representative of atom type (carbon, gray; oxygen, red; hydrogen, white; methyl, light blue). The dotted line in yellow represents the distance, in angstroms, of each of the carbonyl carbons to Thr 1, indicative of potential nucleophilic attack, and to Ser 96, indicative of potential hydrogen bonding. D, in vitro analysis using a 20S proteasome indicates that curcumin inhibits chymotrypsin-like, trypsin-like and PGPH-like activities with IC50 values of 1.85±0.35, 6.23±0.22 and 3.68±0.19 µM, respectively. Ethanol was used as a control (E). Columns, mean of representative independent triplicate experiments; bars, SD.
