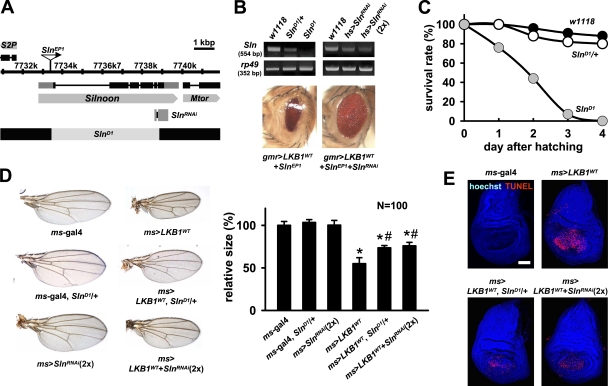
Figure 4.
Sln loss-of-function mutants suppress LKB1-dependent apoptosis. (A) Genomic locus of Sln gene. The deletion and targeting regions of the SlnD1 and the SlnRNAi allele are indicated. (B) RT-PCR analyses of Sln transcript in wild-type (w1118), SlnD1 heterozygous, or SlnD1 homozygous mutant larvae (top left) and in adult flies expressing one copy or two copies of hs-gal4–driven SlnRNAi (top right). rp49 was used as a loading control. Microscopy images of adult eyes expressing gmr-gal4–driven LKB1WT and SlnEP1 without (bottom left) or with (bottom right) SlnRNAi. (C) Survival rates of wild-type (black), SlnD1 heterozygous (white), and SlnD1 homozygous (gray) mutant larvae. (D) Microscopy images of adult wing preparations from the indicated genotypes (left) and the quantification of relative wing sizes (right). Error bars show the SD of 100 independent samples. *, P < 0.05 versus ms-gal4 alone; #, P < 0.05 versus ms>LKB1WT. (E) TUNEL (red) and hoechst (DNA; blue) staining in larval wing discs from the indicated genotypes. Bar, 50 μm.