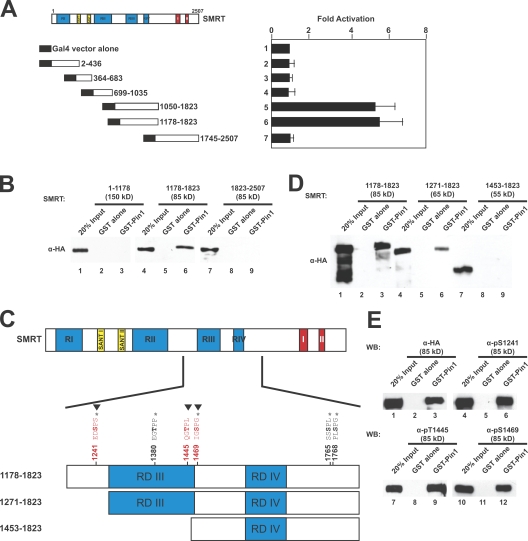
Figure 3.
Mapping the Pin1 interaction domain in SMRT. (A) Yeast two-hybrid mapping of the Pin1 interaction domain. Left, diagram of Gal4-DBD-SMRT fragments used; right, normalized β-galactosidase activity. Amino acids are indicated by numbers. Error bars represent ±SD. (B) GST pull down mapping of Pin1-interacting regions in SMRT. HeLa WCEs expressing the indicated HA-SMRT constructs were subjected to pull downs using GST–Pin1 and immunoblotted with anti-HA antibodies. (C) Schematic diagram of SMRT showing in vivo phosphorylation sites as mapped by mass spectrometry (*), Cdk consensus motifs (▾), and deletion constructs used in Fig. 3 D. Amino acids are indicated by numbers. (D) GST pull downs of N-terminal deletions of HA-SMRT. HeLa WCEs expressing the indicated HA-SMRT constructs were subjected to pull downs using GST–Pin1 and immunoblotted with anti-HA antibodies. (E) Phosphorylated SMRT interacts with Pin1 in vitro. HeLa WCEs expressing HA-SMRT (1,178–1,823) were subjected to pull downs using GST–Pin1 and immunoblotting with the indicated antibodies. WB, Western blot.