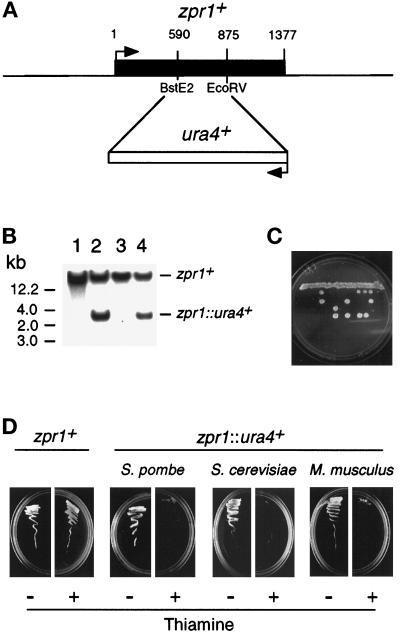
Figure 6.
The zpr1+, gene is essential for cell viability. (A) The S. pombe zpr1+ gene was disrupted by homologous recombination. The structure of the zpr1+ genomic locus and the disrupted gene (zpr1::ura4+) is presented schematically. (B) Southern blot analysis of the diploid yeast transformants. The genomic DNA was restricted with BglII and probed with a random-primed fragment of the zpr1+ genomic locus (2.9-kb XbaI fragment). The wild-type zpr1+ allele was identified in wild-type yeast (strain 480; lane 1). The disrupted zpr1::ura4+ allele (3 kb) was identified in some (lanes 2 and 4) but not all (lane 3) transformants. (C) The heterozygous diploid yeast strain TE630 (zpr1+/zpr1::ura4+) was sporulated, and the tetrads were dissected. The viability of the spores was examined by growth on agar plates supplemented with uracil. (D) The heterozygous (zpr1+/zpr1::ura4+) diploid yeast strain TE630 was transformed with the plasmid pREP41 or the plasmid pREP41-zpr1, selected on minimal agar plates without leucine and uracil, and sporulated, and haploid yeast were selected on minimal media supplemented with adenine. No viable haploid yeast (zpr1::ura4+) were obtained from diploid yeast transformed with pREP41. However, the zpr1 expression vector pREP41-zpr1 complemented the lethal phenotype of the disrupted zpr1+ gene. Complementation was observed in experiments using S. pombe zpr1+, S. cerevisiae ZPR1, and murine ZPR1. Repression of the nmt promoter in the pREP plasmid with thiamine decreased the growth of the complemented ZPR::ura4+ haploid strains, but not the wild-type zpr1+ haploid strain transformed with pREP41-zpr1.