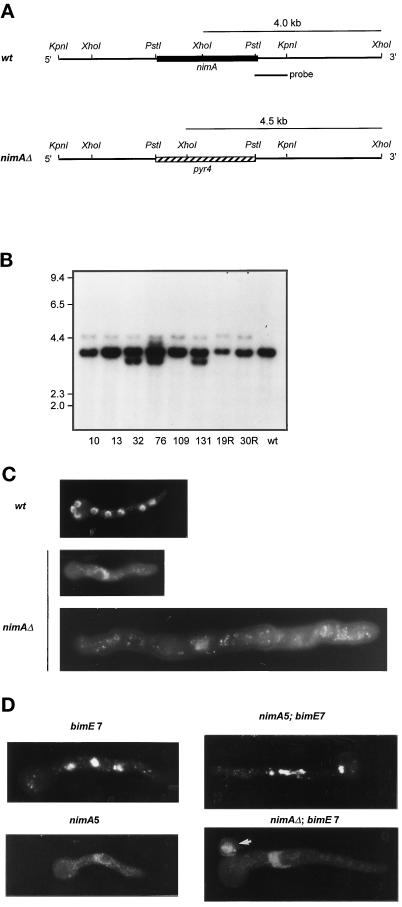
Figure 7.
Targeted nimA deletion. (A) Restriction maps of the nimA locus and nimAΔ locus after targeted deletion. The PstI fragment of nimA was replaced with a PstI fragment containing the pyr4 gene. The linear genomic KpnI fragment containing the pyr4 insert was used for transformation in a wild-type (wt = GR5) and a bimE7APC1 strain. Diagnostic XhoI fragments of 4.0 kb (wt) and 4.5 kb (nimAΔ) when probed with the PstI–KpnI fragment are indicated. (B) Autoradiogram of a Southern blot of XhoI-digested genomic DNA isolated from putative heterokaryons after deletion of nimA probed with the radiolabled PstI–KpnI fragment indicated in A. 10, 13, 32, 76, 109, and 131 are deletions in GR5, and 19R and 30R indicate deletions in a bimE7APC1 strain. Heterokaryons 10, 13, 109, 19R, and 30R all contain both a specific deletion and the wild-type allele. (C) Micrographs of DAPI-stained germlings. The wild-type germling is from an 8-h sample and contains eight interphase nuclei. The nimAΔ germlings are derived from a heterokaryon germinated for 8 and 15 h, respectively, at 32°C. (D) Representative micrographs of DAPI-stained germlings from strains with the indicated pertinent genotypes. Spores were first allowed to germinate at 32°C for 7 h before shifting to 42°C for 4 h. The nimA5 mutant germling was grown at 42°C directly for 7 h. It is to be noted that the single nucleus of the large nimAΔ + bimE7APC1 germling remained at interphase upon shift to 42°C for 4 h, whereas nuclei in the bimE7APC1 or nimA5 + bimE7APC1 mutant germlings all became condensed. The arrow indicates an ungerminated spore carrying the parental genotype (GR5 = pyrg89) derived from the nimAΔ + bimE7APC1 heterokaryon.