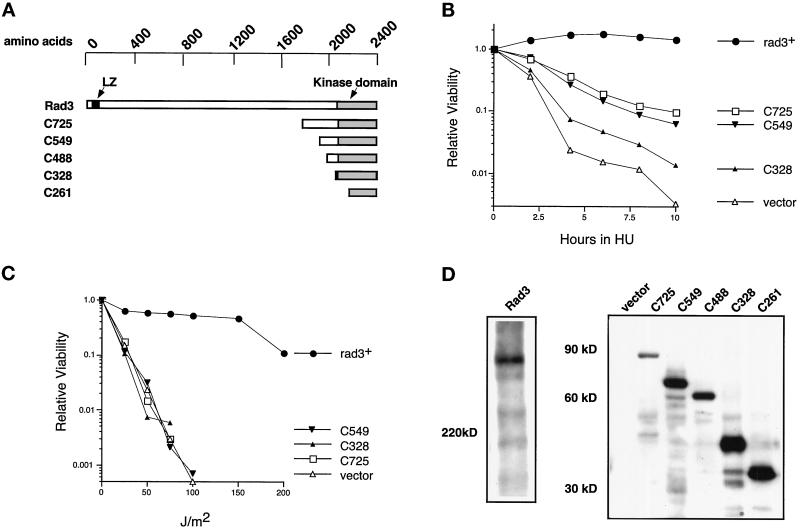
Figure 1.
The isolated Rad3p kinase domain does not complement rad3Δ cells. (A) Rad3p kinase domain fragments are diagrammed approximately to scale. The positions of the kinase domain (shaded) and the leucine zipper motif (black) are indicated. (B) Relative viability of rad3Δ cells (TE890) expressing HA-tagged mutant Rad3 proteins, after indicated periods of incubation in 10 mM HU. SE analysis of these curves indicates that their positions are significant. The plasmids used are rep1-HA-rad3+(pTE541), rep1-HA-rad3-C725 (pTE672), rep1-HA-rad3-C549 (pTE786), rep1-HA-rad3-C328 (pTE784), and rep3x (pTE101). These proteins are overexpressed because their expression is controlled by the full-strength nmt1+ promoter. Note that rep1-HA-rad3+ fully complements the rad3Δ strain, exhibiting wild-type levels of viability in both HU and UV assays (our unpublished results). (C) Relative viability of the same strains after irradiation with the indicated doses of UV. The results shown are the average of two independent experiments. No colonies were recovered at doses at which no points are shown. (D) Expression of full-length Rad3p and Rad3p C-terminal fragments. Left panel, Western blot of total extracts prepared from rad3::ura4+ leu1–32 h− cells (TE890) and cells transformed with rep1-HA-rad3+ (pTE541). Right panel, Western blot of total extracts prepared from the transformants used in B and C, as well as rad3Δ cells (TE890) transformed with rep1-HA-rad3-C261 (pTE785) and rep1-HA-rad3-C488 (pTE787). Both blots were probed with monoclonal antibodies specific for the HA epitope tag (12CA5). Note that direct comparison of levels of expression of full-length to truncations is not feasible, because the different proteins are resolved on different percentage gels and are likely to have different transfer efficiencies. However, we estimate that the truncations are three- to fivefold more abundant than the full-length protein.