SUMMARY
Binding of the HIV envelope to the chemokine coreceptors triggers membrane fusion and signal transduction. The fusion process has been well characterized, yet the role of coreceptor signaling remains elusive. Here we describe a critical function of the chemokine coreceptor signaling in facilitating HIV infection of resting CD4 T cells. We find that static cortical actin in resting T cells represents a restriction, and HIV utilizes the Gαi-dependent signaling from the chemokine coreceptor CXCR4 to activate a cellular actin depolymerizing factor, cofilin, to overcome this restriction. HIV envelope-mediated cofilin activation and actin dynamics are important for a post entry process that leads to viral nuclear localization. Inhibition of HIV-mediated actin rearrangement markedly diminishes viral latent infection of resting T cells. Conversely, induction of active cofilin greatly facilitates it. These findings shed new light on viral exploitation of cellular machinery in resting T cells, where chemokine receptor signaling becomes obligatory.
INTRODUCTION
HIV replication in humans is now thought to utilize two CD4 coreceptors, CCR5 and CXCR4 (Berger et al., 1999). CCR5 is expressed on the activated/memory T cells (Bleul et al., 1997), and appears critical for in vivo susceptibility to infection (Liu et al., 1996). The efficient infection of activated T cells by CCR5 virus is in contrast to the metabolic restrictions of CXCR4 (X4) HIV infection of resting T cells in vitro (Spina et al., 1997), and yet in nearly one half of the HIV-infected population, there is a conversion late in the disease to CXCR4 utilization, corresponding to an accelerated drop in T cell numbers (Fenyo et al., 1988). The unexpected potential for the X4 HIV to expand is borne out by the demonstration of in vivo and ex vivo X4 HIV infection of resting naive T cells (Eckstein et al., 2001; Nishimura et al., 2005; Ostrowski et al., 1999). However, an appreciation of any role of CXCR4, beyond binding and fusion, is lacking.
In vitro, X4 HIV infection of resting T cells is a slow process that can lead to latency with the potential for productive infection (Zack et al., 1990). There is evidence that the ability of HIV to establish latency in resting T cells, albeit slowly, is not purely passive. HIV may alter the cellular environment to facilitate infection and subsequent viral production. For example, expression of Nef in resting T cells can lower the threshold of T cell activation, increasing the likelihood that productive viral replication occurs (Fenard et al., 2005; Schrager and Marsh, 1999; Wu and Marsh, 2001).
During HIV infection, the virus has to undergo entry, uncoating, reverse transcription, and nuclear localization. It is unclear whether these early processes also require modulation of cellular environment by the virus. In particular, at the earliest step, viral binding to its receptors has the potential to trigger critical signaling that may favor viral replication (Cicala et al., 2002). Thus, we tested possible requirements for receptor signaling, both from CD4 and from CXCR4, in viral latent infection of resting T cells. In this article, we report that signal transduction from the chemokine coreceptor CXCR4, but not CD4, is required for HIV latent infection of resting T cells. We also report that the static cortical actin in non-cycling, resting T cells is a post-entry barrier for HIV. To overcome this restriction, the virus utilizes envelope-CXCR4 signaling to activate cofilin, a cellular actin depolymerizing factor critical for actin dynamics and viral nuclear migration. This cofilin restriction is unique to resting T cells, since in transformed or activated T cells cofilin is constitutively active, rendering CXCR4 signaling unnecessary (Alkhatib et al., 1997; Cocchi et al., 1996). Our data uncovered a previously under-appreciated ability of HIV in exploiting the chemokine coreceptor to meet a fundamental need to interact with the actin cytoskeleton (Naghavi et al., 2007). Cofilin activation may contribute to viral infection of resting CD4 T cells in vivo.
RESULTS
Critical Signaling Function of CXCR4 in HIV Infection of Resting CD4 T Cells
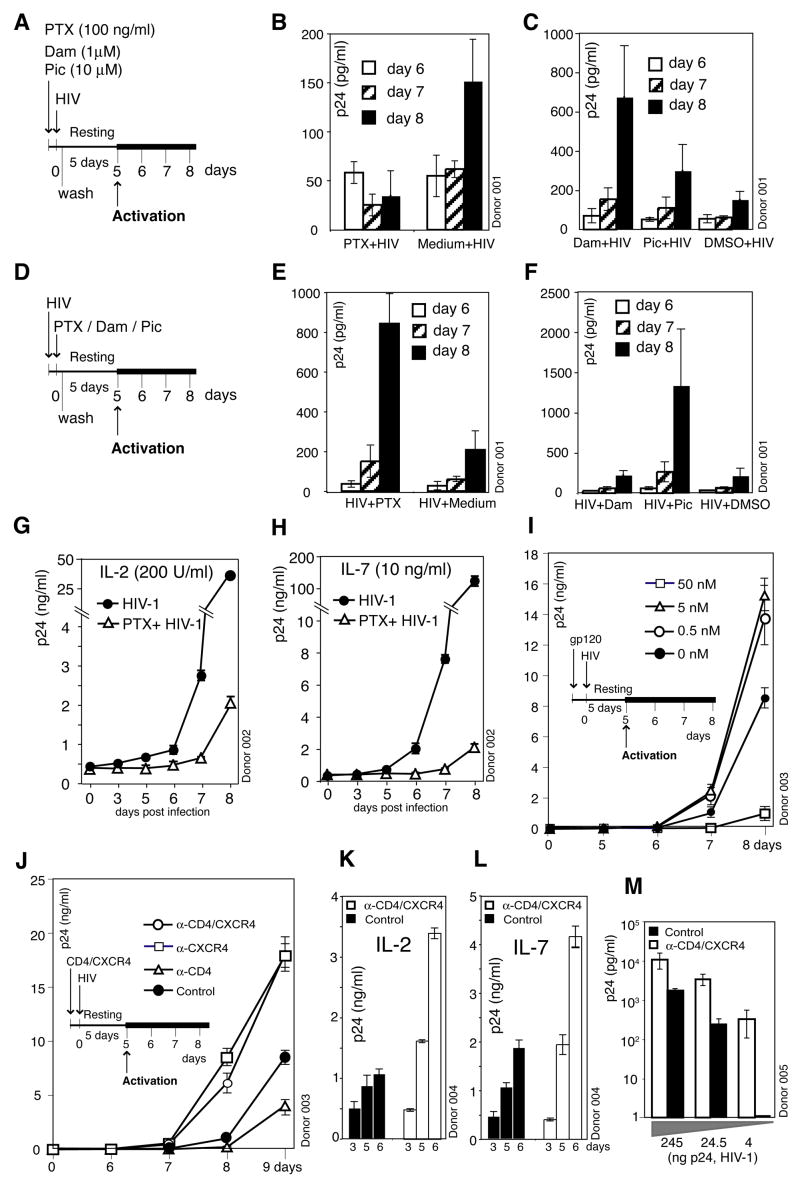
To test possible requirements for receptor signaling, both from CD4 and from CXCR4, in viral infection of resting T cells, we pre-treated cells with inhibitors. The chemokine co-receptor CXCR4 is coupled to G protein, Gαi, which can be uncoupled by treatment with pertussis toxin (PTX). On the other hand, the CD4 receptor is directly coupled with the Src family tyrosine kinase Lck, which can be inhibited by damnacanthal. Resting T cells were briefly treated with these inhibitors, infected with HIV, washed, and incubated for 5 days. During this incubation, productive viral replication does not occur. However, viral replication remains inducible upon T cell activation (Wu and Marsh, 2001). We activated infected cells at day 5 with CD3/CD28 stimulation, and observed inhibition of HIV replication by PTX (Figures 1A, 1B and S1). In contrast, treatment with PTX after infection was not inhibitory (Figures 1D and 1E), excluding the possibility that the PTX inhibition was purely a result of its general toxicity to resting T cells. The slight stimulatory effect of PTX treated after infection could result from the stimulating effect of the B-oligomer of PTX on T cell activity (Gray et al., 1989). In contrast to PTX, damnacanthal did not inhibit viral replication (Figures 1C and 1F). Additionally, piceatannol, an inhibitor targeting the Lck downstream Syk tyrosine kinase ZAP-70, also did not inhibit viral replication (Figures 1C and 1F). These data suggest that signaling from CXCR4, rather than CD4, is important for viral infection of resting T cells. Furthermore, replacement of the viral envelope with the CD4/CXCR4-independent VSV-G envelope avoided the PTX inhibition (Figure S2), demonstrating that the signaling requirement is indeed associated with the HIV envelope. We also tested 8 primary X4 HIV isolates and all of them demonstrated a requirement for CXCR4 signaling (Figure S3). The inhibition of HIV infection by PTX, as we describe here, is unique to resting T cells, since PTX treatment of transformed or activated T cells did not inhibit HIV replication (Figure S4) (Alkhatib et al., 1997; Cocchi et al., 1996).
Figure 1. Requirement of CXCR4 Signaling for HIV Infection of Resting CD4T Cells.
(A to H) Inhibition of HIV infection by pertussis toxin (PTX). Cells were treated with PTX, damnacanthal (Dam), piceatannol (Pic), or medium, DMSO for controls, infected, washed, cultured for 5 days, and then activated with anti-CD3/CD28 beads (A). Shown is viral replication measured by p24 release (B, C). For comparison, cells were infected first, treated with these reagents, washed, and then cultured and activated as above (E, F). (G and H) Cells were also cultured in IL-2 or IL-7 for 4 days, treated with PTX, infected, cultured with cytokines, and then activated as above. (I) Enhancement of HIV infection by gp120. Cells were pre-stimulated with gp120IIIB, infected, incubated and then activated. (J) Enhancement of HIV infection by anti-CD4/CXCR4 beads. Cells were pre-stimulated with anti-CD4/CXCR4, anti-CD4, or anti-CXCR4 beads, infected, incubated and then activated. (K and L) Enhancement of HIV replication by anti-CD4/CXCR4 beads in cytokine-stimulated cells. Cells were cultured in IL-2 or IL-7 for 4 days, pre-stimulated with anti-CD4/CXCR4 beads, infected, washed, and then cultured in cytokines. (M) HIV dosage-dependent enhancement of viral infection by anti-CD4/CXC4 beads. Cells were pre-stimulated with anti-CD4/CXCR4 beads, infected, washed, incubated, and then activated.
Resting T cells circulate between peripheral blood and lymphoid tissues. To determine whether stimulation of resting T cells with lymphatic cytokines can alleviate the viral requirement for CXCR4 signaling (Kreisberg et al., 2006), we cultured resting T cells with IL-2 or IL-7, and observed PTX inhibition of viral replication (Figures 1G and 1H), suggesting that CXCR4 signaling could potentiate infectivity even in certain cytokine-enriched microenvironment such as lymphoid tissues (Figure S5).
To further confirm that binding of HIV envelope to resting T cells triggers positive signaling to facilitate infection, we pre-stimulated cells with the T-tropic gp120. We speculated that this stimulation would provide synergy with the HIV envelope. We found that at low concentrations (0.5 to 5 nM), gp120 enhanced HIV replication (Figures 1I and S6). However, at concentrations above 50 nM, gp120 inhibited HIV, presumably due to competition and down-modulation of CD4 and CXCR4 as previously described (Figure S7). We further tested whether binding of CD4 and CXCR4 was directly responsible for the observed gp120 enhancement, since T-tropic gp120 appears to have high non-specific binding to the cell surface (Babcock et al., 2001). We used beads conjugated with monoclonal antibodies specific for these receptors. Again, we observed enhancement of viral replication by stimulation with anti-CD4/CXCR4 or anti-CXCR4 beads, but not with anti-CD4 beads (Figure 1J). These data are consistent with the inhibition of viral replication by the CXCR4 inhibitor PTX, but not by the CD4 inhibitor damnacanthal or piceatannol (Figures 1B and 1C), suggesting that CXCR4 alone is largely responsible for the delivery of the signal important for viral latent infection. The CD4/CXCR4 enhancement was observed across multiple donors (Figure S8) and also in cytokine-stimulated CD4 T cells (Figures 1K and 1L). The enhancement was most dramatic at low viral dosages (Figure 1M), suggesting that HIV may rely on synergistic engagements of its receptors to fulfil a signaling requirement. The enhancement was neither a result of CD4/CXCR4 stimulation synergizing with that of CD3/CD28 (Figure S9) nor an effect of gp120-induced T cell activation (Figure S10).
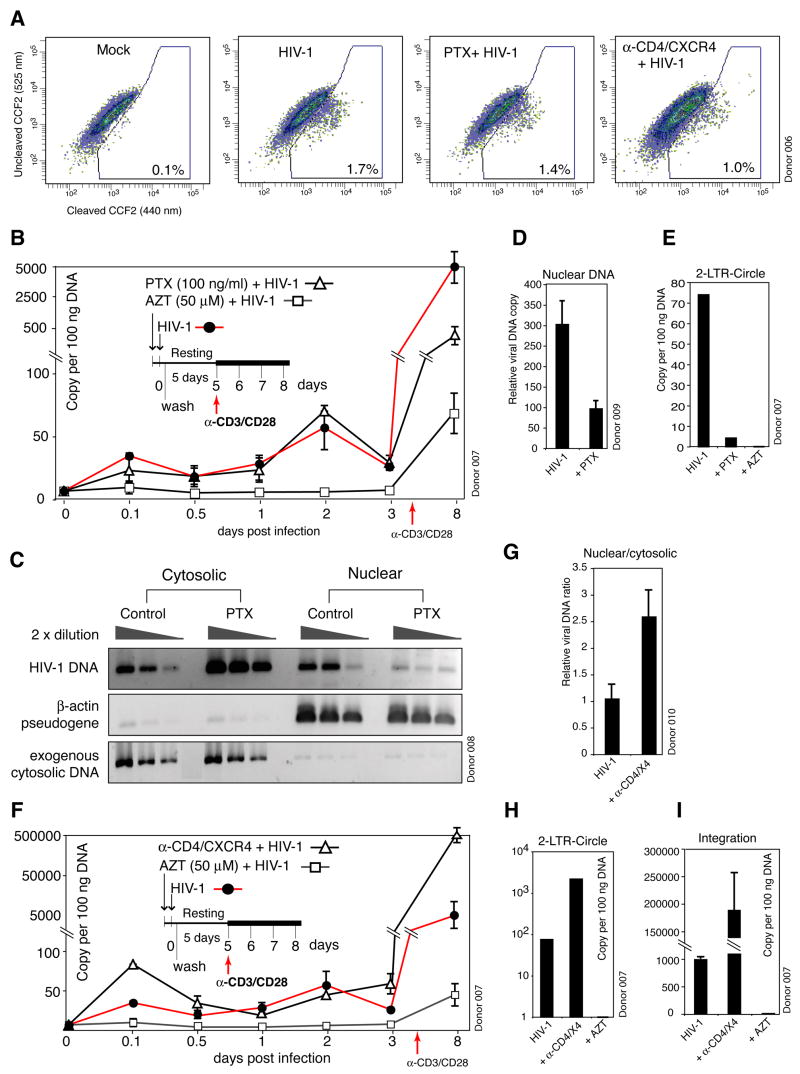
To identify possible viral processes that are sensitive to PTX and thus require CXCR4 signaling, we first examined the effects of PTX treatment on viral receptors and receptor-mediated fusion. PTX treatment did not affect surface CD4/CXCR4 expression (Figure S11), or prevent fusion, although there was a slight decrease (1.7% to 1.4%, Figure 2A). At a low multiplicity of infection (0.02 TCID50 per cell, measured on Rev-CEM) (Wu et al., 2007), viral DNA synthesis peaked at 2 days (Zhou et al., 2005) and PTX did not affect the peak level of viral DNA (Figure 2B). Along with the fusion experiments, these results indicate that entry or reverse transcription is not the process displaying PTX sensitivity. We then examined viral DNA nuclear targeting. A sizable fraction of viral DNA migrated into the nucleus as early as 2 hours (Figure 2C), immediately following signaling events triggered by gp120. In contrast, in PTX-treated cells, viral DNA nuclear migration was decreased (Figure 2C). This was confirmed again by the examination of the viral nuclear DNA at day 3. Although the total viral DNA was similar, the nuclear DNA was less in PTX-treated cells (Figures 2B and 2D). This is consistent with a measurable decrease of 2-LTR circles (Figure 2E).
Figure 2. Molecular Characterization of Viral Processes that Require CXCR4 Signaling in Resting CD4 T Cells.
(A) Effects of PTX and anti-CD4/CXCR4 stimulation on viral fusion. Cells were treated with PTX, or with anti-CD4/CXCR4 beads, infected with HIV(BlaM-Vpr) or not infected (Mock), washed, and then loaded with CCF2 for flow cytometry. (B) Effects of PTX on viral DNA synthesis. Cells were treated with PTX and infected. Viral DNA synthesis was measured by real-time PCR (three measurements with standard error). AZT was used as a control. (C, D) PTX Inhibition of viral DNA nuclear localization. Cells were treated with PTX, infected for 2 hours, washed, lysed, and then fractionated into cytoplasmic and nuclear fractions. DNA was amplified for HIV late DNA, nuclear β-actin pseudogene or an exogenously added plasmid as fractionation controls (C). Nuclear DNA in cells at day 3 was also measured by real-time PCR (D). (E) is a real-time PCR measurement of 2-LTR circles in cells at day 8. (F) Effects of CD4/CXCR4 stimulation on viral DNA synthesis. Cells were stimulated with anti-CD4/CXCR4 beads and infected. Viral DNA synthesis was measured by real-time PCR. (G) Enhancement of viral DNA nuclear localization by CD4/CXCR4 stimulation. Cells were stimulated with anti-CD4/CXCR4 beads, infected for 2 hours, and then fractionated into cytoplasmic and nuclear fractions. Viral DNA was measured by real-time PCR. The average ratio of viral nuclear to cytoplasmic DNA in unstimulated control was arbitrarily assigned as “1”. (H and I) is a real-time PCR measurement of 2-LTR circles (H) and viral DNA integration (I) in cells at day 8.
Since CD4/CXCR4 stimulation enhanced viral replication (Figure 1J), we also followed viral processes in stimulated cells. We observed that early, transient viral DNA synthesis (2 hours) was enhanced by CD4/CXCR4 stimulation (Figure 2F). This enhancement did not result from an increase in fusion (Figure 2A), but may occur at a post entry step such as uncoating, reverse transcription, or nuclear import. Indeed, viral nuclear migration was enhanced in CD4/CXCR4-stimulated cells (Figure 2G). Interestingly, in CD4/CXCR4-stimulated cells, the kinetics of viral DNA synthesis and decay was altered. Although the viral DNA synthesis at day 2 was similar, there was no decrease of viral DNA at day 3 in CD4/CXCR4-stimulated cells. Fractionation of the nuclear DNA at day 3 also revealed that viral nuclear DNA was increased in the stimulated cells (data not shown). The increased amount of nuclear DNA also correlated with higher levels of 2-LTR circles and viral integration (Figures 2H and 2I). These data demonstrate that CD4/CXCR4-stimulation promotes viral DNA nuclear localization, and further suggest that nuclear migration may subvert the viral DNA decay process. In unstimulated resting T cells, the viral DNA synthesized at day 2 was largely diminished after day 3 (Figure 2F). This may result from a lack of nuclear migration signaling in the absence of continual CXCR4 stimulation after removal of the input virus. We conclude that in addition to the functions of viral binding and entry, CXCR4 serves to contribute signaling that is critical for viral DNA nuclear migration. It is reasonable that this results in an inherent increase in HIV DNA stability in resting T cells.
HIV Envelope-CXCR4 Signaling Promotes Cortical Actin Dynamics
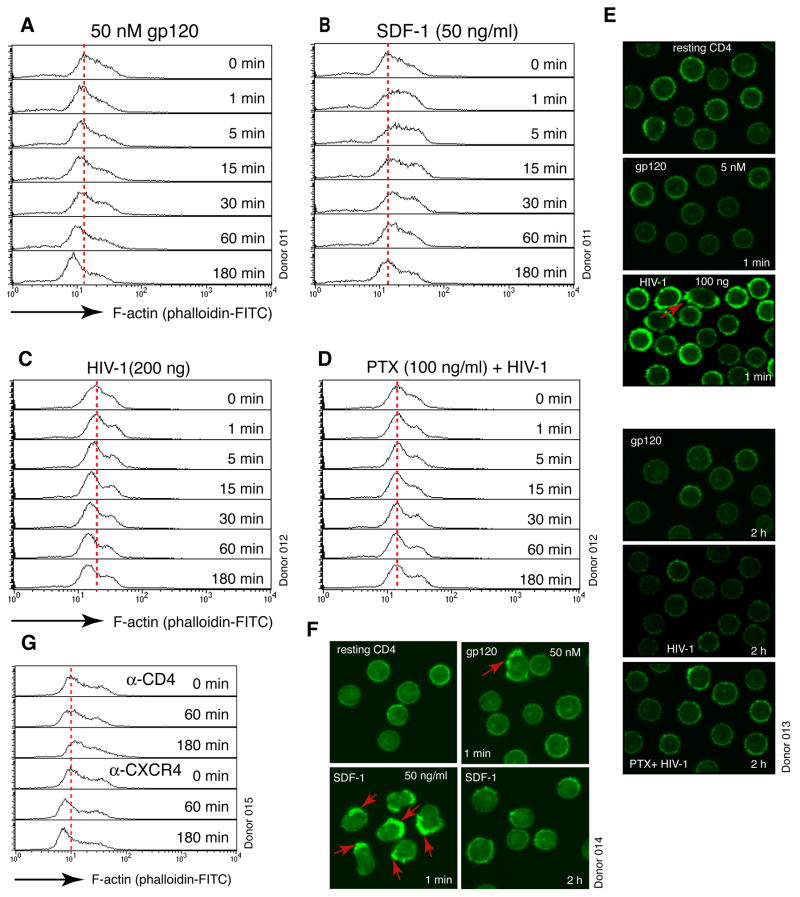
In T cells, signaling through CXCR4 leads to chemotactic T cell motility and actin polymerization (Sotsios et al., 1999). Given that gp120 has the capacity to trigger signaling through CXCR4, we examined the possible effects of gp120 on the actin cytoskeleton of resting T cells. We observed that treatment of resting T cells with gp120, HIV or anti-CXCR4 beads triggers the polymerization and depolymerization of the cortical actin filaments (F-actin) (Figures 3A, 3C, 3G and S12 to S16). Although localized cortical actin polymerization by gp120 or HIV was occasionally seen within 1 minute of stimulation (Balabanian et al., 2004) (Figures 3E, 3F and S12 to S16), actin depolymerization was the most consistent observation, and it usually occurred after 5 minutes and became more pronounced at 1 to 2 hours (Figures 3A and 3C). As a control, stimulation with SDF-1, the natural ligand of CXCR4, also triggers rapid, polarized cortical actin polymerization (Balabanian et al., 2004) (Figures 3B and 3F). However, for HIV or gp120, at the low dosages we used, it was generally not capable of triggering such a drastic, global cytoskeletal rearrangement as SDF-1. The gp120-mediated actin change was observed in multiple donors and across a range of gp120 concentrations tested, from 5 pM to 50 nM (Figures 3A and S12), and was seen with multiple primary gp120 and HIV isolates (Figure S16). Replacement of gp120 with the VSV-G envelope abolished the ability of HIV to trigger actin changes (Figure S17). Furthermore, depletion of Nef (Campbell et al., 2004) from the virion particles did not prevent gp120-mediated actin depolymerization (Figure S18). These data confirm that gp120 alone was largely responsible for modulating F-actin. Other virion factors such as Nef likely act at the post fusion steps (Tobiume et al., 2003). The decrease in F-actin in response to HIV or gp120 stimulation did not result from a global loss of the actin protein (Figure S19). Cellular fractionation of F-actin and globular actin monomer (G-actin) further confirmed changes in the ratio of F-actin to G-actin in response to gp120 treatment (Figure S20).
Figure 3. HIV Envelope-CXCR4 Signaling Promotes Cortical Actin Dynamics.
(A, B) Actin changes mediated by gp120 or SDF-1. Cells were treated with gp120IIIB or SDF-1, fixed, permeabilized, and then stained with FITC-phalloidin for flow cytometry. (C, D) HIV-mediated actin depolymerization was dependent on PTX-sensitive Gαi signaling. Cells were not treated (C) or treated with PTX (D) for 2 hours, infected, and then stained with FITC-phalloidin. (E, F) Confocal microscopy of FITC-phalloidin staining of cells treated with gp120IIIB, SDF-1, HIV or PTX plus HIV. The full time courses of these treatments are shown in Figures S12 to S14. Red arrows indicate localized actin polymerization. (G) Actin depolymerization triggered by stimulation of CXCR4. Cells were stimulated with either anti-CD4 or anti-CXCR4-IMag particles, and stained with FITC-phalloidin.
Actin polymerization induced by SDF-1 is regulated through the activation of G proteins, particularly the PTX-sensitive Gαi (Sotsios et al., 1999). Similarly, gp120-mediated actin depolymerization was also found to be mediated through Gαi (Figures 3C and 3D). The fact that PTX inhibited both viral replication and actin change suggests that promoting actin dynamics could be one of the essential functions of gp120-CXCR4 signaling in priming viral infection and nuclear localization (Figures 1B, 3D and 2C). It is possible that in the absence of cell cycle or chemotactic stimulation, the cortical actin in resting T cells could be relatively static and represent a restriction for the virus. As such, increasing cortical actin activity might be required to achieve intracellular migration of HIV. It has been known that actin-based motility is critical for the movement of a variety of intracellular pathogens such as Listeria monocytogenes, Shigella flexneri, and vaccinia virus. Most of these pathogens encode molecular mimics such as ActA, VirG and A36R to promote actin dynamics (Cameron et al., 2000).
Viral Requirement for Actin Reorganization in Resting CD4 T Cells
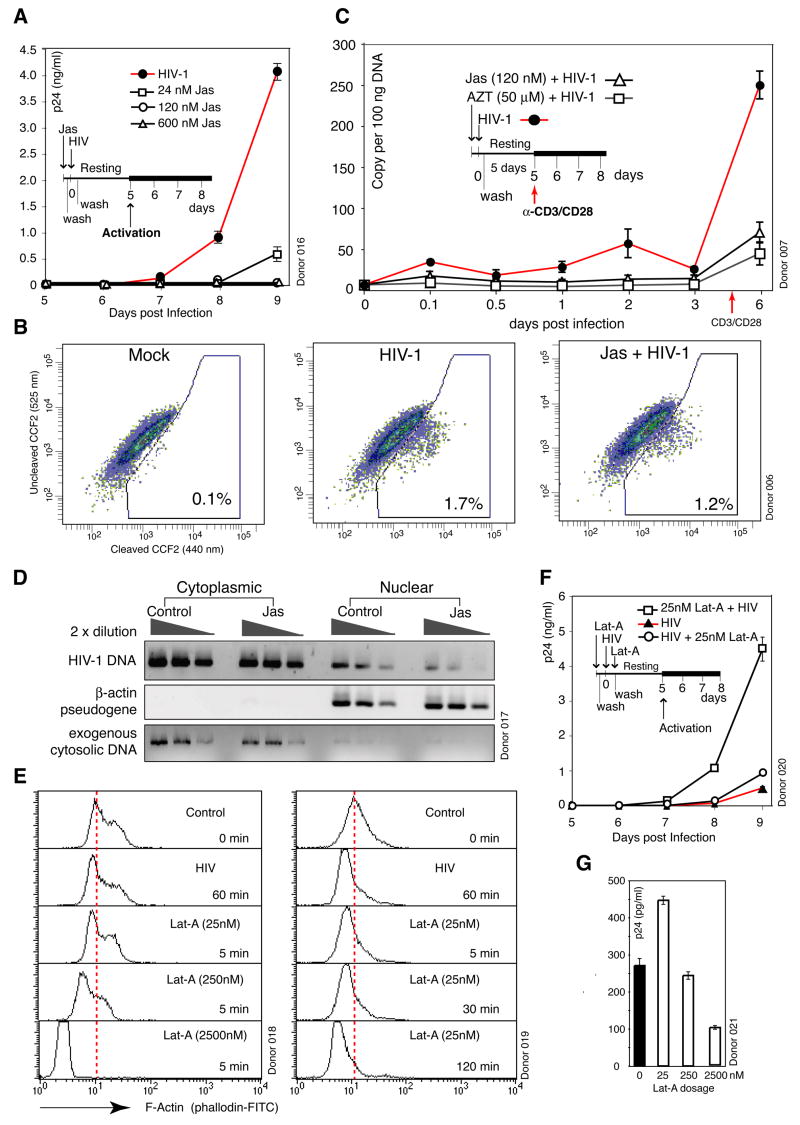
To further address the requirement of actin reorganization for infection of resting T cells, we used an F-actin stabilizing agent, jasplakinolide (Jas), to interfere with gp120-induced actin change. Jas binds to F-actin irreversibly and prevents depolymerization of actin filaments. We speculated that Jas could affect HIV latent infection through direct inhibition of actin change. We observed complete inhibition of HIV replication by Jas at 120 nM or higher concentrations, and partial inhibition at 24 nM (Figure 4A). Jas inhibition was observed across multiple donors, although there were donor-dependent variations in the degree of inhibition at low Jas dosages (data not shown).
Figure 4. Viral Requirement for Actin Activity in HIV Infection of Resting CD4 T Cells.
(A) Jas inhibition of HIV infection. Cells were treated with Jas for 2 hours, washed, infected, washed, cultured, and then activated with anti-CD3/CD28 beads to initiate viral replication. (B) Effects of Jas on viral fusion. Cells were treated with 120 nM Jas, infected with HIV(BlaM-Vpr), washed, and then loaded with CCF2 for measuring fusion. (C) Effects of Jas on viral DNA synthesis. Cells were treated with 120 nM Jas for 1 hour and infected. Viral DNA synthesis was measured by real-time PCR. AZT was used as a control. (D) Inhibition of viral DNA nuclear localization by 120 nM Jas. Cells were treated with 120 nM Jas, infected, washed, lysed, and then fractionated into cytoplasmic and nuclear fractions as in Figure 2C. (E) Dosage and time-dependent actin depolymerization by Lat-A. Cells were treated with Lat-A for 5 min (left panel), or with 25 nM Lat-A for various times (right panel), and stained with FITC-phalloidin for flow cytometry. Untreated and HIV-infected cells were used as controls. (F) Enhancement of HIV infection by Lat-A. Cells were treated with 25 nM Lat-A for 5 minutes, washed, infected, washed, cultured, and then activated. As a control, cells were also infected first, washed, treated with Lat-A for 5 minutes, washed, and then incubated and activated identically. (G) Dosage-dependent effects of Lat-A on HIV infection. Cells were treated with Lat-A for 5 min at different dosages, infected, washed, incubated, and then activated.
The inhibition of viral replication by 120 nM and 24 nM Jas was neither a result of inhibition on T cell activation (Figures S21 to S23) nor an effect of altered CD4/CXCR4 receptor distribution (Figure S24) that prevented fusion (Figure 4B, only a slight decrease from 1.7% to 1.2%). A time course analysis of viral DNA synthesis found that 120 nM Jas persistently inhibited viral DNA synthesis (Figure 4C). DNA fractionation further demonstrated that the viral nuclear DNA was diminished (Figure 4D). These data suggest that binding of Jas irreversibly to F-actin may competitively inhibit viral association with F-actin, and thus, inhibited viral reverse transcription and nuclear migration. These findings are consistent with an earlier proposal that establishment of the viral reverse transcription complex is dependent on contact with actin microfilaments (Bukrinskaya et al., 1998).
The Jas inhibition on actin rearrangement could be obviated by T cell activation since similar 120 nM Jas treatment of transformed or activated T cells (Figures S25 and S26A) did not inhibit HIV replication. It is likely that at 120 nM, Jas does not inhibit the cell cycle or cell cycle-mediated actin remodelling; thus, new actin filaments generated may be free of Jas. At a higher dosage (3 μM), however, when the cell cycle was arrested, Jas did inhibit HIV replication in cycling T cells (Figure S26B). Additionally, CD4/CXCR4 pre-stimulation of resting T cells also completely abolished the inhibition by 120 nM Jas (Figure S27), likely as a result of increased actin dynamics, prompted by CD4/CXCR4 stimulation.
To further confirm that the static actin cytoskeleton in unstimulated resting T cells constitutes a restriction for HIV, we used another actin modulator, Latrunculin A (Lat-A), which has been shown to specifically induce actin depolymerization through reversible binding to G-actin. We speculated that induction of actin depolymerization by Lat-A may enhance HIV replication if the cortical actin is relatively static and serves as a barrier. To test this hypothesis, we first titrated Lat-A at various dosages, and found that at high dosages (2.5 μM to 250 nM), Lat-A induced dramatic actin depolymerization, whereas at a lower dosage (25 nM) it induced actin depolymerization to an extent similar to that induced by HIV (Figure 4E). Thus, we pre-treated resting T cells with 25 nM Lat-A and consistently observed enhancement of HIV replication in all donors examined (Figure 4F). We also tested Lat-A at higher dosages where Lat-A induced actin depolymerization was much greater than the physiological depolymerization induced by HIV (Figure 4E). Interestingly, at 250 nM, we observed donor-dependent variations from enhancement to inhibition (data not shown). At 2.5 μM, however, Lat-A inhibited viral replication in all donors (Figure 4G). These data imply that excessive, non-physiological depolymerization may affect T cell function or viral activity. Nevertheless, our data conclusively demonstrate that irreversible stabilization of actin by Jas inhibits HIV replication, whereas slight depolymerization of actin by Lat-A enhances viral replication. Collectively, these findings suggest that the static actin cytoskeleton in resting T cells represents a barrier that needs to be dynamically reorganized, and HIV may exploit the CXCR4 signaling pathway to fulfil this requirement.
HIV Envelope-CXCR4 Signaling Triggers Activation of Cofilin to Promote Actin Dynamics
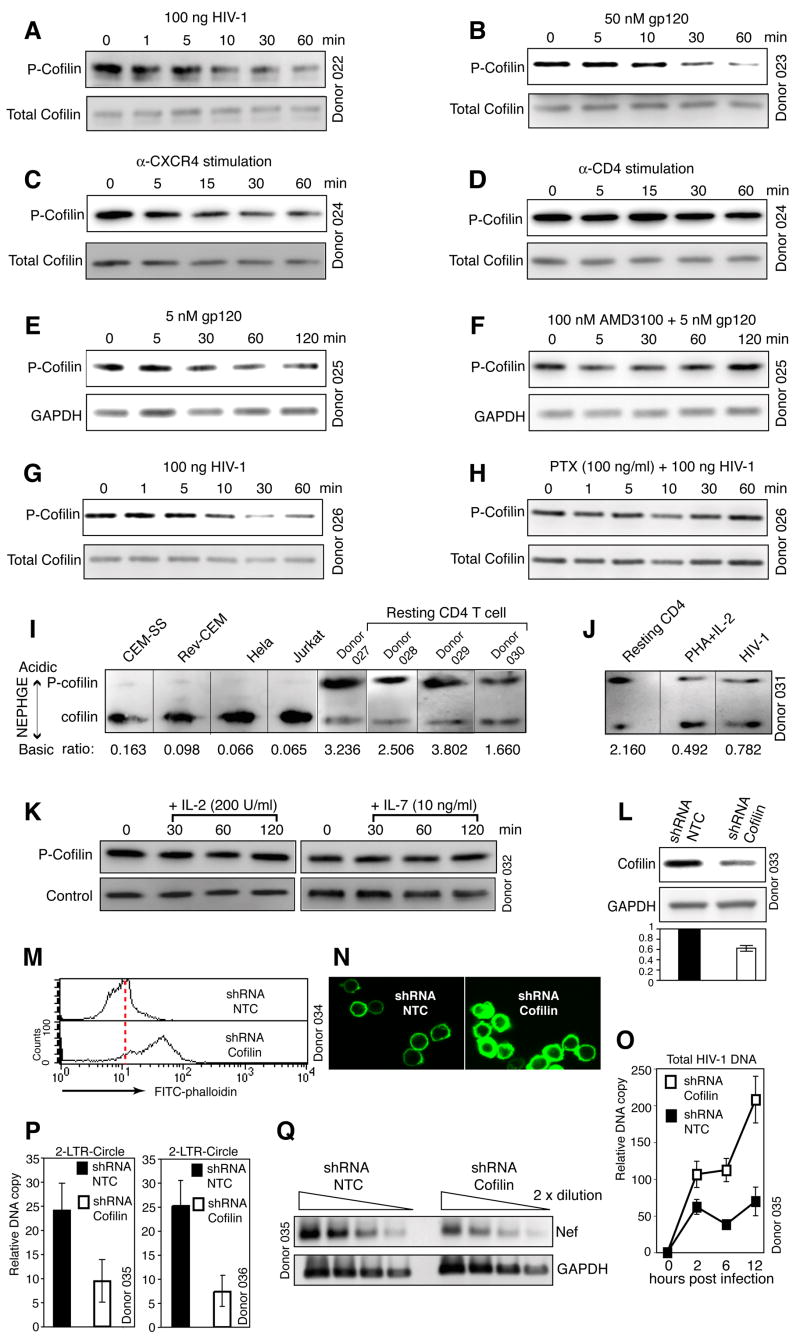
In eukaryotic cells, assembly and disassembly of actin filaments are modulated primarily by the actin-depolymerizing factor (ADF)/cofilin family of proteins (Bamburg et al., 1980). The most important physiological function of cofilin is to sever and depolymerize actin filaments, thereby promoting actin dynamics (Lappalainen and Drubin, 1997). Cofilin activity is regulated by phosphorylation at serine 3, which prevents the association of cofilin with actin (Arber et al., 1998), whereas dephosphorylation by phosphatases activates cofilin (Ambach et al., 2000; Gohla et al., 2005; Niwa et al., 2002). Given that HIV envelope-CXCR4 signaling mediates actin depolymerization, we examined cofilin activity in resting T cells and in cells infected with HIV. We observed that in resting T cells, cofilin was largely inactivated by phosphorylation, and upon HIV infection, cofilin was activated by dephosphorylation within minutes (Figure 5A). HIV-induced cofilin activation in resting T cells was seen in all donors examined and with multiple primary isolates of HIV (Figure S28A). Moreover, the kinetics correlated well with HIV-induced actin depolymerization, with strong cofilin activation and actin depolymerization occurring between 1 to 2 hours (Figure 3C). Occasionally, we observed quick actin polymerization within 1 min (Figure S13), and this correlated with a transient increase in cofilin phosphorylation within 1 min, and was then followed by cofilin dephosphorylation and activation (data not shown). Cofilin activation was further observed by treatment of resting T cells with gp120 (Figure 5B) or anti-CXCR4 beads (Figure 5C), but not with anti-CD4 beads (Figure 5D), demonstrating that engagement of CXCR4 is sufficient to activate cofilin. Furthermore, pre-treatment of resting CD4 T cells with a highly specific CXCR4 antagonist, AMD3100 (Gerlach et al., 2001), or PTX completely inhibited cofilin activation by HIV (Figures 5E to 5H and S28B). This is consistent with PTX inhibition of actin depolymerization by HIV (Figure 3D). Based on these data, we conclude that HIV envelope binding to CXCR4 triggers PTX-sensitive activation of cofilin and its association with F-actin (Figure S29) to promote actin dynamics.
Figure 5. HIV Envelope-CXCR4 Signaling Triggers PTX-sensitive Activation of Cofilin.
(A to D) Activation of cofilin by HIV, gp120 or CXCR4 stimulation. Cells were treated with HIV (A), gp120 IIIB (B), anti-CXCR4 beads (C), or anti-CD4 beads (D), and then analyzed by Western blot using an anti-phospho-cofilin antibody (P-cofilin). The blots were stripped and reprobed with an antibody against total cofilin. (E, F) AMD3100 inhibits gp120-triggered cofilin activation. Cells were not treated (E) or treated with AMD3100 (F), stimulated with gp120, and then analyzed by Western blot. (G, H) PTX-sensitive activation of cofilin by HIV. Cells were not treated (G) or treated with PTX (H), infected, and then analyzed by Western blot. (I) Constitutive activation of cofilin in transformed cell lines. P-cofilin (upper band) and active cofilin (lower band) were separated and detected by NEPHGE-Western blot using an anti-cofilin antibody. The relative ratio of P-cofilin to active cofilin is indicated at the bottom. (J) Cofilin activation induced by PHA plus IL-2 treatment of resting T cells. HIV-infected cells were used as a control. Cells were analyzed by NEPHGE-Western blot. (K) IL-2 or IL-7 stimulation does not activate cofilin. Resting T cells were stimulated with IL-2 or IL-7, and then analyzed by Western blot. (L to Q) shRNA knockdown of cofilin in CD4 T cells. knockdown procedure is in Figure S31. Cells carrying cofilin shRNA or a control shRNA (NTC) were analyzed by Western blot using an anti-human cofilin antibody or an anti-human GAPDH antibody (L). (M, N) Suppression of cofilin expression leads to an accumulation of F-actin as measured by FITC-phalloidin staining and analyses with flow cytometry or confocal microscopy. (O) is a real-time PCR measurement of viral DNA synthesis in knockdown cells. (P) is a real-time PCR measurement of HIV 2-LTR circles in knockdown cells at 12 hours (left panel) and 24 hours (right panel) post infection. (Q) is a RT-PCR analysis of the same cells at 12 hours, measuring HIV nef and the cellular GAPDH transcripts.
Contrary to resting T cells, activated or transformed T cells do not require CXCR4 signaling to support viral replication. It is possible that in these cells cofilin is differently regulated (Samstag et al., 1994). Indeed, when the ratio of phospho-cofilin to active cofilin was compared, transformed or activated T cells predominantly carry dephosphorylated, active cofilin (Figures 5I and 5J), suggesting that T cell activation promotes cofilin activity, rendering CXCR4 signaling unnecessary for HIV. Unlike T cell activation, stimulation of resting T cells with cytokines such as IL-2 and IL-7 did not activate cofilin (Figure 5K), and consistently, the presence of these cytokines did not alleviate the requirement for CXCR4 signaling (Figures 1G and 1H). We conclude that cofilin activation is a unique event occurring in HIV infection of resting T cells or cells exposed to certain cytokines.
To further determine the requirement for cofilin activity in HIV-mediated actin change and viral infection, we suppressed cofilin expression to approximately two-thirds in CD4 T cells utilizing small interfering RNA (siRNA) (Figures 5L and S31E). The cofilin knockdown cells demonstrated significantly higher levels of cortical actin filaments (Figures 5M, 5N, S31F, and S31G), in agreement with our results showing that induction of cofilin activity by gp120, an inverse of cofilin suppression, led to the depolymerization of F-actin (Figures 3A, 3C, 5A and 5B). The cofilin knockdown cells also displayed a diminished capacity to support HIV replication (Figure S31H). However, given that cofilin is a fundamental protein involved in multiple cellular processes, the inhibition on viral replication may not be viral specific. Thus, we directly examined viral early processes following infection. We found a higher level of total viral DNA early following infection of the cofilin knockdown cells (Figure 5O), suggesting that fusion, entry, and reverse transcription were not adversely affected. However, viral 2-LTR circles were decreased by approximately 2.5- and 3-fold at 12 and 24 hours (Figure 5P), suggesting a decrease of viral DNA nuclear migration in the cofilin knockdown cells. If normalized to total viral DNA, the cofilin knockdown cells had an approximate 90% reduction in nuclear viral DNA at 12 hours, in comparison with the control. Quantification of viral early transcripts at 6 and 12 hours also showed that the nef transcript was decreased (Figure 5Q), a confirmation of diminished viral nuclear DNA templates available in the cofilin knockdown cells. Based on these data we conclude that suppressing cofilin activity led to an aberrant accumulation of cortical actin and an impediment to viral nuclear migration. This is consistent with a viral requirement for CXCR4 signaling and cofilin activation to mediate cortical actin change for nuclear migration. The mechanism for the early increase in total viral DNA in cofilin knockdown cells is unknown, but could result from possible stimulatory effects of F-actin to viral transcriptional process (Bukrinskaya et al., 1998; De et al., 1993; Hottiger et al., 1995).
Cofilin Activation Promotes HIV Latent Infection of Resting CD4 T Cells
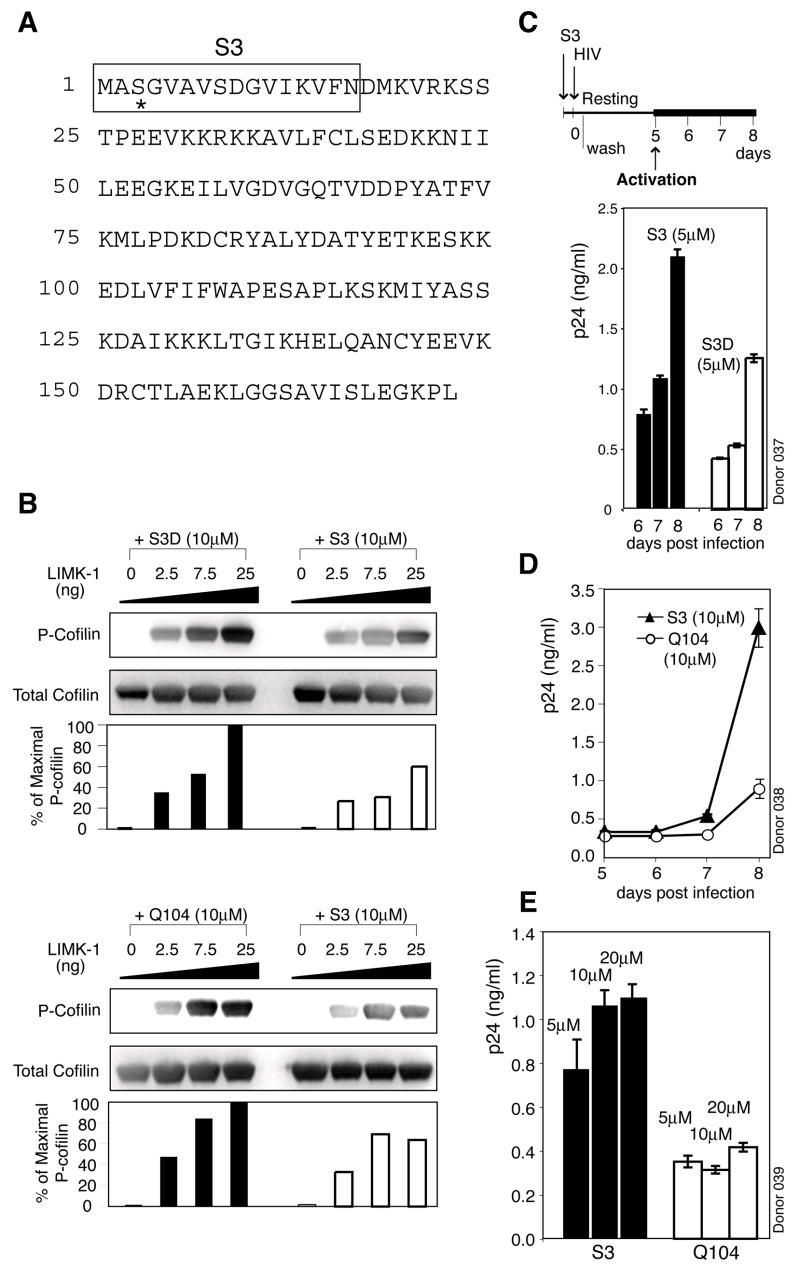
As an alternative approach to confirm the role of cofilin in HIV infection, we also pursued modulation of cofilin activity by altering upstream signals. In resting T cells, cofilin is inhibited by phosphorylation at serine-3. The inhibition is maintained through the basal activity of the LIM family protein kinases (LIMK), for which ADF/cofilin proteins are the only known substrates (Arber et al., 1998). To date, there is no known specific inhibitor for LIM kinases. To test whether activation of cofilin is directly involved in viral latent infection, we used a synthetic peptide to compete with cofilin for LIMK1 to inhibit cofilin phosphorylation. This peptide, S3, carries the N-terminal 16 residues, including serine 3 of human cofilin (Nishita et al., 2002) (Figure 6A). In in vitro LIMK1 kinase assay, we observed that S3 directly inhibited cofilin phosphorylation by LIMK1 (Figure 6B). We next tested whether activation of cofilin by S3 would enhance viral latent infection. We observed enhancement of viral replication by S3 (Figures 6C to 6E), and this enhancement did not result from enhancement on T cell activation through CD3/CD28 stimulation (Figure S32).
Figure 6. Induction of Cofilin Activation by S3 Peptide Promotes Viral Latent Infection of Resting CD4 T Cells.
(A) Human non-muscle cofilin protein sequence. The sequence of S3 is shown in the boxed region and serine 3 is denoted with asterisk. (B) S3 activates cofilin through competitive inhibition of LIMK1. In vitro LIMK kinase assay was performed using purified LIMK1 and GST-tagged recombinant human cofilin in the presence of S3, S3D or Q104. Cofilin phosphorylation was analyzed by Western blot. (C to E) Enhancement of viral replication by S3. Cells were treated with S3, S3D or Q104 for 2 hours, infected, washed, cultured, and then activated to initiate viral replication. (E) shows a S3 dosage-dependent enhancement of viral replication at day 8.
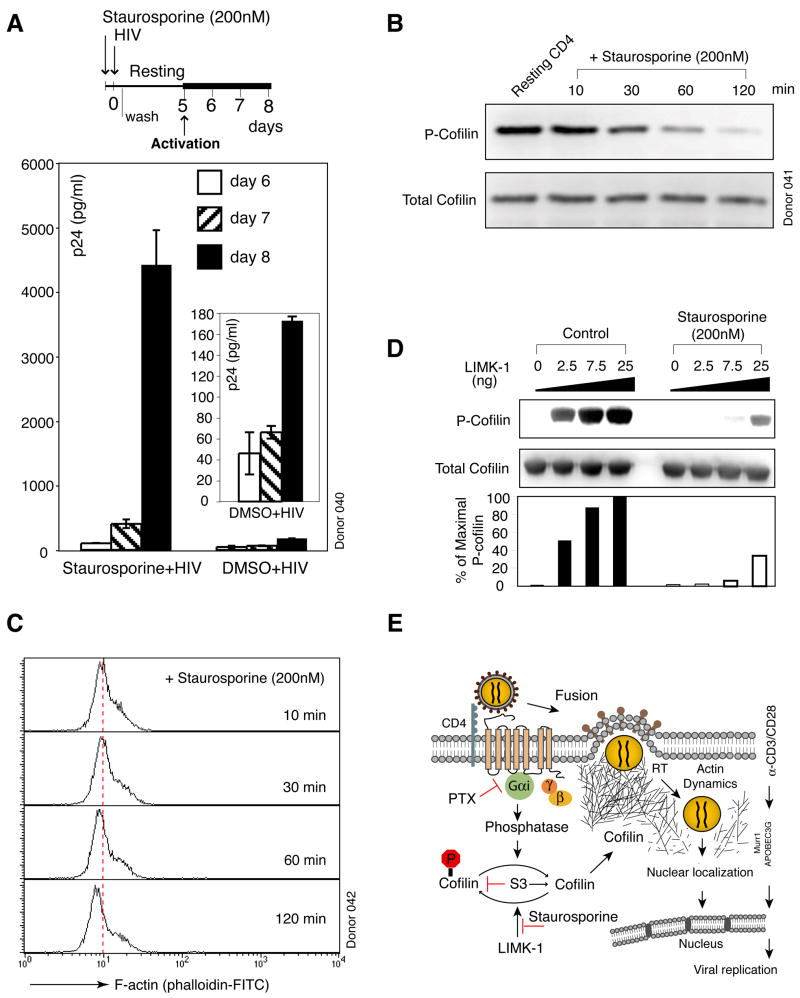
In addition to S3, we have also tested multiple kinase inhibitors to probe HIV-mediated signaling in resting T cells (Figure 1). Of these, we observed that staurosporine, a general serine/threonine kinase inhibitor, yielded an unexpected result. While this inhibitor largely shuts down T cell activation (Kubbies et al., 1989) (Figure S33), the transient treatment of resting CD4 T cells with staurosporine prior to infection, but not afterwards (Figure S34), led to a dramatic enhancement in HIV replication (Figure 7A). Given the broad impact of staurosporine on cell signal transduction, we felt compelled to examine possible effects of this general kinase inhibitor on cofilin activation. We found that the brief treatment of resting CD4 T cells with staurosporine led to gradual dephosphorylation and activation of cofilin in the absence of any stimulation (Figure 7B). With the activation of cofilin, actin depolymerization was also observed in staurosporine-treated cells (Figure 7C). These features remarkably resemble those initiated by HIV infection of resting T cells (Figures 3C and 5A). Within the cofilin pathway, staurosporine appears to act on LIMK1-mediated cofilin phosphorylation directly, since the kinase activity assay showed that staurosporine strongly decreased the phosphorylation of cofilin by LIMK1 (Figure 7D). Thus, the unexpected ability of staurosporine to activate cofilin and enhance viral replication, through transient exposure before infection, appears consistent with the identified role of cofilin in HIV infection.
Figure 7. Induction of Cofilin Activation by Staurosporine Promotes Viral Latent Infection of Resting CD4 T Cells.
(A) Enhancement of viral replication by staurosporine. Cells were treated with staurosporine for 2 hours, infected, washed, incubated, and then activated to initiate viral replication. (B) Staurosporine induces cofilin activation in resting T cells. Cells were treated with staurosporine and analyzed by Western blot. (C) Staurosporine induces actin depolymerization in resting T cells. Cells were treated with staurosporine and stained with FITC-phalloidin for flow cytometry. (D) Staurosporine induces cofilin activation by direct inhibition of LIMK1. In vitro LIMK kinase assay was performed in the presence or absence of staurosporine. (E) Model of gp120-CXCR4 signaling in mediating cofilin activation and HIV latent infection. Binding of gp120 to CXCR4 triggers fusion and signal transduction that leads to cofilin activation. Following fusion, the viral preintegration complex is directly anchored onto F-actin to facilitate reverse transcription (Bukrinskaya et al., 1998). Subsequent actin activity mediated by cofilin activation increases cortical actin dynamics and actin treadmilling, which promote the movement of the viral preintegration complex toward the centre of the cell.
Collectively, our data demonstrate that activation of cofilin is one of the most important events in HIV latent infection of resting T cells. Because the chemokine receptor signaling is diverse, we do not exclude other possible functions of the chemokine receptor signaling in facilitating HIV infection in the various cellular states found in vivo.
DISCUSSION
Here we report that HIV-mediated CXCR4 signaling actively takes part in an early post entry step, specifically, nuclear localization, for the latent infection of resting T cells. We also identify the targeted molecular activity of CXCR4 signaling, the activation of cofilin. While it is straightforward to envision that the cortical actin in resting cells is a barrier to nuclear import, the successful delivery of the preintegration complex to the nucleus likely depends on other cytoskeletal functions. Indeed, another cytoskeletal factor, moesin, has been shown recently to directly influence retroviral infectivity (Naghavi et al., 2007), and important interactions between the viral components and the cellular cytoskeleton have been previously documented; retroviruses may use the actin cytoskeleton to facilitate transfer of viral genome from the peripheral regions of the cell to the microtubule network for reverse transcription and post entry migration (Bukrinskaya et al., 1998; McDonald et al., 2002; Naghavi et al., 2005). The viral nucleocapsid (Wilk et al., 1999), reverse transcriptase (Hottiger et al., 1995), and integrase (Turlure et al., 2004) have been known to directly interact with actin, suggesting possible anchorage of the preintegration complex onto the actin network. Our results also suggest that the actin cytoskeleton in resting CD4 T cells may not simply be a barrier, as excessive depolymerization of actin by Lat-A inhibits viral replication (Figures 4E and 4G). It is reasonable that the direct interaction of the virus with the cortical actin mass could facilitate uncoating, reverse transcription, or allow HIV to gain access to the perinuclear and nuclear region (Figure 7E).
We identify cofilin as a critical factor required for HIV latent infection of resting CD4 T cells. Importantly, cofilin is different from previously identified restriction factors, such as Murr1 (Ganesh et al., 2003) or the low molecular mass APOBEC3G (LMM A3G) (Chiu et al., 2005). While knockdown of Murr1 or LMM A3G in resting T cells can lead to productive viral replication, activation of cofilin by gp120, S3, or staurosporine did not. Activation of cofilin appears to be a naturally evolved function of gp120 by viral selection of the CXCR4 chemokine receptor.
In the human immune system, cofilin regulates actin dynamics and is involved in T cell migration and activation (Eibert et al., 2004; Nishita et al., 2002). It has been known that a genetic defect affecting actin dynamics due to deficiency of WASP causes immunodeficiency (Symons et al., 1996). It was also predicted that abnormalities in cofilin or its regulatory molecules could cause T cell-mediated immunodeficiency (Bamburg and Wiggan, 2002). The demonstration of cofilin as a critical target of the viral envelope suggests that this biochemical pathway in CXCR4 T cells could be aberrantly altered in HIV infected individuals. Indeed, we have extended our study into HIV-infected patients. Our preliminary data suggest that in their CD4 T cells, cofilin activity is aberrantly up-regulated (data not shown).
Cofilin is the primary molecule regulating cortical actin dynamics (Lappalainen and Drubin, 1997). In cycling cells, cofilin is constitutively active to facilitate constant remodelling of the actin cytoskeleton. In contrast, in resting CD4 T cells cofilin is largely inactive, implying a less dynamic cortical actin that may represent a realistic barrier for the virus. Activation of cofilin is an apparent solution, and HIV exploits the chemokine receptor to meet this fundamental need at the earliest step of its life cycle.
EXPERIMENTAL PROCEDURES
Additional details for methods are provided in the Supplemental Data.
Treatment and Infection of Resting CD4 T Cells
Resting CD4 T cells were purified from peripheral blood as previously described (Wu and Marsh, 2001). Cells were treated with pertussis toxin (Sigma), jasplakinolide (Invitrogen), staurosporine (Biomol) for 2 hours or latrunculin A (Biomol) for 5 minutes, and then infected with HIV. For HIV infection, unless specified, 103.5 to 104.5 TCID50 units (measured on Rev-CEM) (Wu et al., 2007) were used to infect 106 cells. Following infection, cells were washed 2 to 3 times. Cells were treated with synthetic peptide S3, or the control peptide S3D, Q104 for 1 to 2 hours, infected with HIV.
FITC-Phalloidin Staining of F-actin and Flow Cytometry
Cells were treated with gp120 IIIB (Microbix Biosystems) from 1 minute to 2 hours, fixed, permeabilized, and then stained with 5 μl of 0.3 mM FITC-labelled phalloidin for 30 minutes at 4°C. After washing, cells were resuspended in 1% paraformaldehyde and analyzed on a FACSCalibur (Becton Dickinson).
Measurement of Cofilin and Phospho-cofilin by Western Blot
Cells were lysed in NuPAGE LDS Sample Buffer (Invitrogen), separated by SDS-PAGE, transferred onto nitrocellulose membranes (Invitrogen), and then blocked for 30 minutes with Starting Block buffer (Pierce). The blots were incubated with either a rabbit anti-cofilin antibody (1:1000 dilution) (Cell Signaling) or a rabbit anti-phospho-cofilin (ser3) antibody (1:1000 dilution) (Cell Signaling). The blots were washed, then incubated with goat anti-rabbit horseradish peroxidase-conjugated antibodies (1:1000 dilution) (KPL). The blots were developed with SuperSignal West Femto Maximum Sensitivity Substrate (Pierce).
In vitro LIMK Kinase Assay
LIMK1 kinase assay was performed using purified LIMK1 and GST-tagged recombinant human cofilin (Upstate Biotechnologies). Briefly, recombinant cofilin was incubated in 1 X Kinase reaction buffer in the presence or absence of staurosporine or the S3, S3D, Q104 peptides. LIMK1 was serially diluted in dilution buffer, and then added into the reaction along with the ATP buffer. The reaction was incubated for 15 minutes at 30°C.
Supplementary Material
Acknowledgments
We thank GMU Student Health Center; Department of Transfusion Medicine, NIH; V. Chandhoke and C. Bailey for blood donation; NIH AIDS Research & Reference Reagent Program for reagents; H. A. Nash of NIMH/NIH; K. Jeang of NIAID/NIH; D. Rekosh of UVA; L. Liotta, E. Petricoin and D. Cox for comments; T. Meckel and T. Jin of NIAID/NIH for confocal microscopy and imaging analyses; K. L. Holmes of NIAID/NIH for fusion assay; A. Biancotto and L. Margolis of NICHHD/NIH for tonsillar T cells; Y. Samstag for protocols, and A. Wu for digital graphic design. A. Yoder was supported by NDSE Fellowship. This work was supported by George Mason University, the Intramural Program of the NIMH/NIH, and in part by the Public Health Service grant AI069981 from NIAID to Y.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alkhatib G, Locati M, Kennedy PE, Murphy PM, Berger EA. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology. 1997;234:340–348. doi: 10.1006/viro.1997.8673. [DOI] [PubMed] [Google Scholar]
- Ambach A, Saunus J, Konstandin M, Wesselborg S, Meuer SC, Samstag Y. The serine phosphatases PP1 and PP2A associate with and activate the actin-binding protein cofilin in human T lymphocytes. Eur J Immunol. 2000;30:3422–3431. doi: 10.1002/1521-4141(2000012)30:12<3422::AID-IMMU3422>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- Babcock GJ, Mirzabekov T, Wojtowicz W, Sodroski J. Ligand binding characteristics of CXCR4 incorporated into paramagnetic proteoliposomes. J Biol Chem. 2001;276:38433–38440. doi: 10.1074/jbc.M106229200. [DOI] [PubMed] [Google Scholar]
- Balabanian K, Harriague J, Decrion C, Lagane B, Shorte S, Baleux F, Virelizier JL, Arenzana-Seisdedos F, Chakrabarti LA. CXCR4-tropic HIV-1 envelope glycoprotein functions as a viral chemokine in unstimulated primary CD4+ T lymphocytes. J Immunol. 2004;173:7150–7160. doi: 10.4049/jimmunol.173.12.7150. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, Harris HE, Weeds AG. Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett. 1980;121:178–182. doi: 10.1016/0014-5793(80)81292-0. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, Wiggan OP. ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 2002;12:598–605. doi: 10.1016/s0962-8924(02)02404-2. [DOI] [PubMed] [Google Scholar]
- Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinskaya A, Brichacek B, Mann A, Stevenson M. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J Exp Med. 1998;188:2113–2125. doi: 10.1084/jem.188.11.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron LA, Giardini PA, Soo FS, Theriot JA. Secrets of actin-based motility revealed by a bacterial pathogen. Nat Rev Mol Cell Biol. 2000;1:110–119. doi: 10.1038/35040061. [DOI] [PubMed] [Google Scholar]
- Campbell EM, Nunez R, Hope TJ. Disruption of the actin cytoskeleton can complement the ability of Nef to enhance human immunodeficiency virus type 1 infectivity. J Virol. 2004;78:5745–5755. doi: 10.1128/JVI.78.11.5745-5755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- Cicala C, Arthos J, Selig SM, Dennis G, Jr, Hosack DA, Van Ryk D, Spangler ML, Steenbeke TD, Khazanie P, Gupta N, et al. HIV envelope induces a cascade of cell signals in non-proliferating target cells that favor virus replication. Proc Natl Acad Sci USA. 2002;99:9380–9385. doi: 10.1073/pnas.142287999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchi F, DeVico AL, Garzino-Demo A, Cara A, Gallo RC, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- De BP, Burdsall AL, Banerjee AK. Role of cellular actin in human parainfluenza virus type 3 genome transcription. J Biol Chem. 1993;268:5703–5710. [PubMed] [Google Scholar]
- Eckstein DA, Penn ML, Korin YD, Scripture-Adams DD, Zack JA, Kreisberg JF, Roederer M, Sherman MP, Chin PS, Goldsmith MA. HIV-1 Actively Replicates in Naive CD4(+) T Cells Residing within Human Lymphoid Tissues. Immunity. 2001;15:671–682. doi: 10.1016/s1074-7613(01)00217-5. [DOI] [PubMed] [Google Scholar]
- Eibert SM, Lee KH, Pipkorn R, Sester U, Wabnitz GH, Giese T, Meuer SC, Samstag Y. Cofilin peptide homologs interfere with immunological synapse formation and T cell activation. Proc Natl Acad Sci USA. 2004;101:1957–1962. doi: 10.1073/pnas.0308282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenard D, Yonemoto W, de Noronha C, Cavrois M, Williams SA, Greene WC. Nef is physically recruited into the immunological synapse and potentiates T cell activation early after TCR engagement. J Immunol. 2005;175:6050–6057. doi: 10.4049/jimmunol.175.9.6050. [DOI] [PubMed] [Google Scholar]
- Fenyo EM, Morfeldt-Manson L, Chiodi F, Lind B, von Gegerfelt A, Albert J, Olausson E, Asjo B. Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J Virol. 1988;62:4414–4419. doi: 10.1128/jvi.62.11.4414-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh L, Burstein E, Guha-Niyogi A, Louder MK, Mascola JR, Klomp LW, Wijmenga C, Duckett CS, Nabel GJ. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature. 2003;426:853–857. doi: 10.1038/nature02171. [DOI] [PubMed] [Google Scholar]
- Gerlach LO, Skerlj RT, Bridger GJ, Schwartz TW. Molecular interactions of cyclam and bicyclam non-peptide antagonists with the CXCR4 chemokine receptor. J Biol Chem. 2001;276:14153–14160. doi: 10.1074/jbc.M010429200. [DOI] [PubMed] [Google Scholar]
- Gohla A, Birkenfeld J, Bokoch GM. Chronophin, a novel HAD-type serine protein phosphatase, regulates cofilin-dependent actin dynamics. Nat Cell Biol. 2005;7:21–29. doi: 10.1038/ncb1201. [DOI] [PubMed] [Google Scholar]
- Gray LS, Huber KS, Gray MC, Hewlett EL, Engelhard VH. Pertussis toxin effects on T lymphocytes are mediated through CD3 and not by pertussis toxin catalyzed modification of a G protein. J Immunol. 1989;142:1631–1638. [PubMed] [Google Scholar]
- Hottiger M, Gramatikoff K, Georgiev O, Chaponnier C, Schaffner W, Hubscher U. The large subunit of HIV-1 reverse transcriptase interacts with beta-actin. Nucleic Acids Res. 1995;23:736–741. doi: 10.1093/nar/23.5.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisberg JF, Yonemoto W, Greene WC. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J Exp Med. 2006;203:865–870. doi: 10.1084/jem.20051856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbies M, Goller B, Russmann E, Stockinger H, Scheuer W. Complex Ca2+ flux inhibition as primary mechanism of staurosporine-induced impairment of T cell activation. Eur J Immunol. 1989;19:1393–1398. doi: 10.1002/eji.1830190807. [DOI] [PubMed] [Google Scholar]
- Lappalainen P, Drubin DG. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naghavi MH, Hatziioannou T, Gao G, Goff SP. Overexpression of fasciculation and elongation protein zeta-1 (FEZ1) induces a post-entry block to retroviruses in cultured cells. Genes Dev. 2005;19:1105–1115. doi: 10.1101/gad.1290005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naghavi MH, Valente S, Hatziioannou T, de Los Santos K, Wen Y, Mott C, Gundersen GG, Goff SP. Moesin regulates stable microtubule formation and limits retroviral infection in cultured cells. Embo J. 2007;26:41–52. doi: 10.1038/sj.emboj.7601475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Brown CR, Mattapallil JJ, Igarashi T, Buckler-White A, Lafont BA, Hirsch VM, Roederer M, Martin MA. Resting naive CD4+ T cells are massively infected and eliminated by X4-tropic simian-human immunodeficiency viruses in macaques. Proc Natl Acad Sci USA. 2005;102:8000–8005. doi: 10.1073/pnas.0503233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishita M, Aizawa H, Mizuno K. Stromal cell-derived factor 1alpha activates LIM kinase 1 and induces cofilin phosphorylation for T-cell chemotaxis. Mole Cell Biol. 2002;22:774–783. doi: 10.1128/MCB.22.3.774-783.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- Ostrowski MA, Chun TW, Justement SJ, Motola I, Spinelli MA, Adelsberger J, Ehler LA, Mizell SB, Hallahan CW, Fauci AS. Both memory and CD45RA+/CD62L+ naive CD4(+) T cells are infected in human immunodeficiency virus type 1-infected individuals. J Virol. 1999;73:6430–6435. doi: 10.1128/jvi.73.8.6430-6435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstag Y, Eckerskorn C, Wesselborg S, Henning S, Wallich R, Meuer SC. Costimulatory signals for human T-cell activation induce nuclear translocation of pp19/cofilin. Proc Natl Acad Sci USA. 1994;91:4494–4498. doi: 10.1073/pnas.91.10.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrager JA, Marsh JW. HIV-1 Nef increases T cell activation in a stimulus-dependent manner. Proc Natl Acad Sci USA. 1999;96:8167–8172. doi: 10.1073/pnas.96.14.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotsios Y, Whittaker GC, Westwick J, Ward SG. The CXC chemokine stromal cell-derived factor activates a Gi-coupled phosphoinositide 3-kinase in T lymphocytes. J Immun. 1999;163:5954–5963. [PubMed] [Google Scholar]
- Spina CA, Prince HE, Richman DD. Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro. J Clin Invest. 1997;99:1774–1785. doi: 10.1172/JCI119342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M, Derry JM, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Tobiume M, Lineberger JE, Lundquist CA, Miller MD, Aiken C. Nef does not affect the efficiency of human immunodeficiency virus type 1 fusion with target cells. J Virol. 2003;77:10645–10650. doi: 10.1128/JVI.77.19.10645-10650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlure F, Devroe E, Silver PA, Engelman A. Human cell proteins and human immunodeficiency virus DNA integration. Front Biosci. 2004;9:3187–3208. doi: 10.2741/1472. [DOI] [PubMed] [Google Scholar]
- Wilk T, Gowen B, Fuller SD. Actin associates with the nucleocapsid domain of the human immunodeficiency virus Gag polyprotein. J Virol. 1999;73:1931–1940. doi: 10.1128/jvi.73.3.1931-1940.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Beddall MH, Marsh JW. Rev-dependent indicator T cell line. Curr HIV Res. 2007;5:395–403. doi: 10.2174/157016207781024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Marsh JW. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science. 2001;293:1503–1506. doi: 10.1126/science.1061548. [DOI] [PubMed] [Google Scholar]
- Zack JA, Arrigo SJ, Weitsman SR, Go AS, Haislip A, Chen IS. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhang H, Siliciano JD, Siliciano RF. Kinetics of human immunodeficiency virus type 1 decay following entry into resting CD4+ T cells. J Virol. 2005;79:2199–2210. doi: 10.1128/JVI.79.4.2199-2210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.