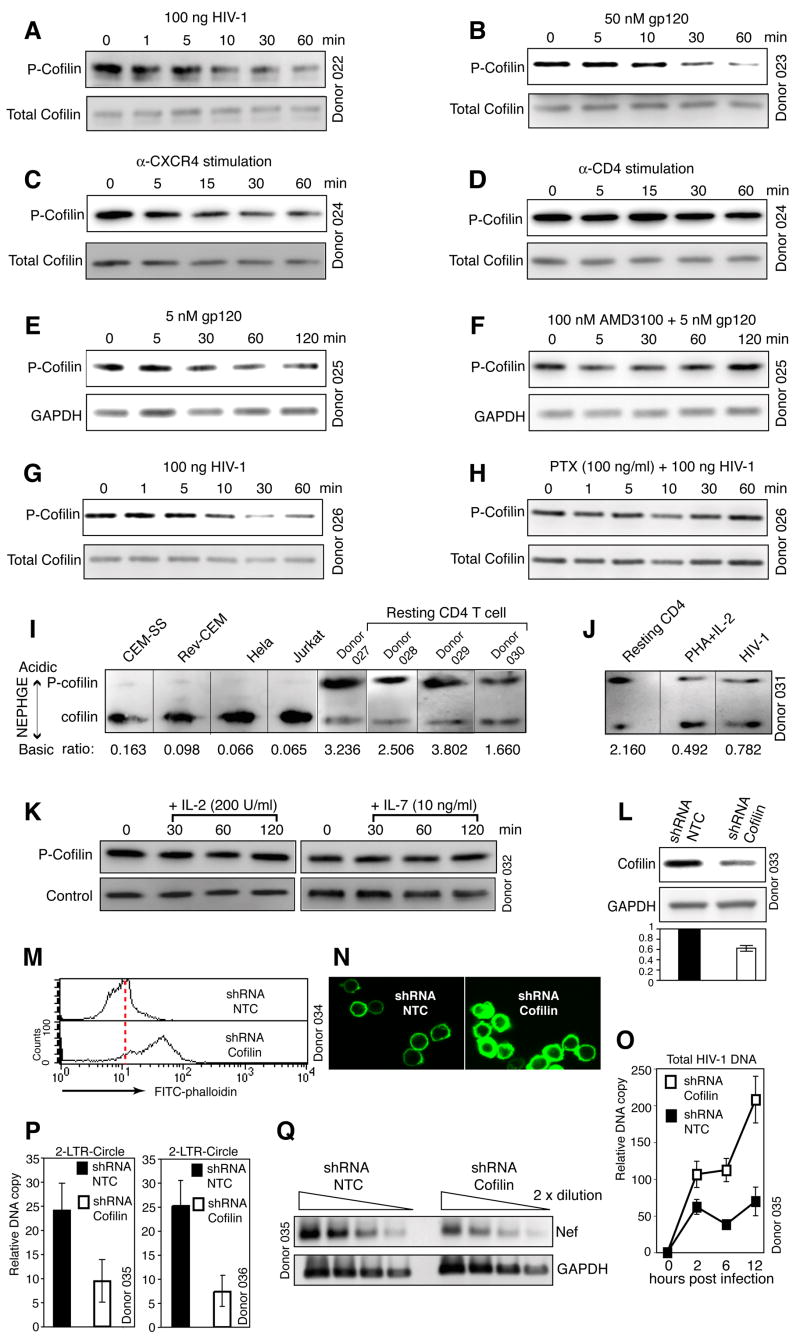
Figure 5. HIV Envelope-CXCR4 Signaling Triggers PTX-sensitive Activation of Cofilin.
(A to D) Activation of cofilin by HIV, gp120 or CXCR4 stimulation. Cells were treated with HIV (A), gp120 IIIB (B), anti-CXCR4 beads (C), or anti-CD4 beads (D), and then analyzed by Western blot using an anti-phospho-cofilin antibody (P-cofilin). The blots were stripped and reprobed with an antibody against total cofilin. (E, F) AMD3100 inhibits gp120-triggered cofilin activation. Cells were not treated (E) or treated with AMD3100 (F), stimulated with gp120, and then analyzed by Western blot. (G, H) PTX-sensitive activation of cofilin by HIV. Cells were not treated (G) or treated with PTX (H), infected, and then analyzed by Western blot. (I) Constitutive activation of cofilin in transformed cell lines. P-cofilin (upper band) and active cofilin (lower band) were separated and detected by NEPHGE-Western blot using an anti-cofilin antibody. The relative ratio of P-cofilin to active cofilin is indicated at the bottom. (J) Cofilin activation induced by PHA plus IL-2 treatment of resting T cells. HIV-infected cells were used as a control. Cells were analyzed by NEPHGE-Western blot. (K) IL-2 or IL-7 stimulation does not activate cofilin. Resting T cells were stimulated with IL-2 or IL-7, and then analyzed by Western blot. (L to Q) shRNA knockdown of cofilin in CD4 T cells. knockdown procedure is in Figure S31. Cells carrying cofilin shRNA or a control shRNA (NTC) were analyzed by Western blot using an anti-human cofilin antibody or an anti-human GAPDH antibody (L). (M, N) Suppression of cofilin expression leads to an accumulation of F-actin as measured by FITC-phalloidin staining and analyses with flow cytometry or confocal microscopy. (O) is a real-time PCR measurement of viral DNA synthesis in knockdown cells. (P) is a real-time PCR measurement of HIV 2-LTR circles in knockdown cells at 12 hours (left panel) and 24 hours (right panel) post infection. (Q) is a RT-PCR analysis of the same cells at 12 hours, measuring HIV nef and the cellular GAPDH transcripts.