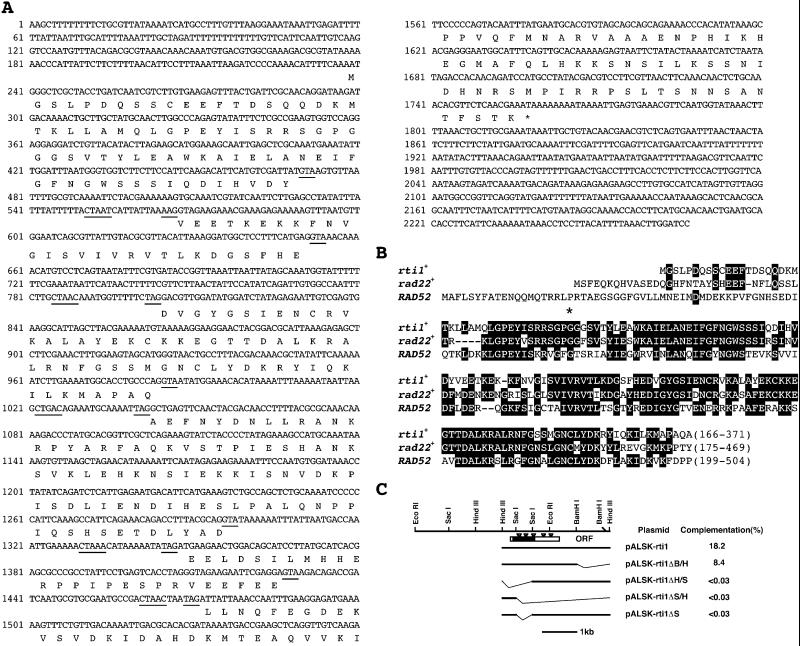
Figure 1.
Structure of rti1+. (A) Nucleotide sequence of rti1+ and the deduced amino acid sequence of the putative gene product. Underlines within introns indicate consensus splicing sequences. Splice junctions were determined by comparing the genomic and cDNA sequences of rti1+. The predicted 371-amino-acid sequence is indicated in single-letter code. (B) Amino acid alignment of the predicted Rti1 protein with Rad22 and Rad52. The aa residues identical among them are boxed. The asterisk indicates the position of mutation in the rad22-H6 gene. In this mutant allele, there is a G to A conversion at nucleotide 441 resulting in an amino acid change from glycine to aspartic acid at amino acid 110. (C) Organization and deletion analysis of rti1+. The activity of variously deleted rti1+ gene was expressed as percent complementation of the thermosensitivity of rad22-H6 cells. The filled box indicates the highly homologous region, and inverted triangles indicate the positions of introns. Thin lines are deleted regions.