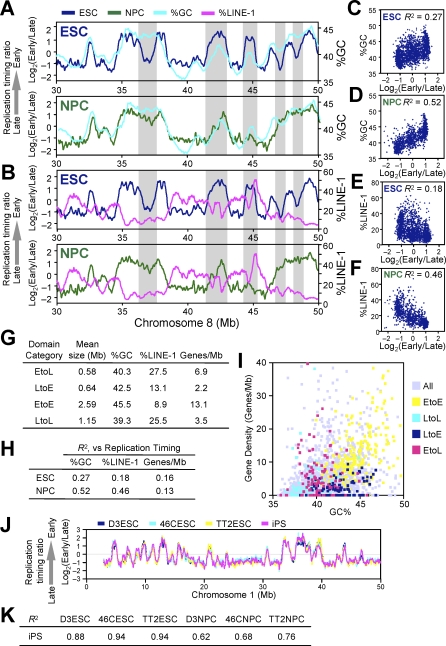
Figure 4. Domain Consolidation Aligns Replication Timing to GC/LINE-1 Content but Not Gene Density.
(A and B) Loess-smoothed replication-timing profiles of ESCs (blue) and NPCs (green) were overlaid with moving averages of 500-kb windows of GC (A) and LINE-1 (B) content for a segment on Chromosome 8. Grey highlighted areas show regions where differentiation aligns replication timing to GC/LINE-1 content.
(C–F) Average replication-timing ratios of replication domains in ESCs (C and E) and NPCs (D and F) were plotted against their GC content (C and D) and LINE-1 content (E and F). Pearson's R 2 values are shown.
(G) EtoL and LtoE domains have an unusual combination of GC/LINE-1 content and gene density. Domains with the 5% greatest replication-timing changes were defined as EtoL and LtoE, whereas those with the least changes (lowest 20%) that maintained replication timing ratio of above 0.5 or below −0.5 were defined as EtoE and LtoL, respectively. Genes/Mb refers to RefSeq gene density.
(H) Correlation of gene density and replication timing at the level of domains in ESCs and NPCs. Pearson R 2 values are shown.
(I) Scatter plot of GC content and gene density shows that EtoE, LtoL, LtoE, and EtoL domains are generally GC rich/gene rich, GC poor/gene poor, GC rich/gene poor, and GC poor/gene rich, respectively.
(J and K) Replication domain structure of iPS cells matches that of ESCs both by visual inspection (J) and high Pearson R 2 values for pair-wise comparisons of iPS to ESCs (K).