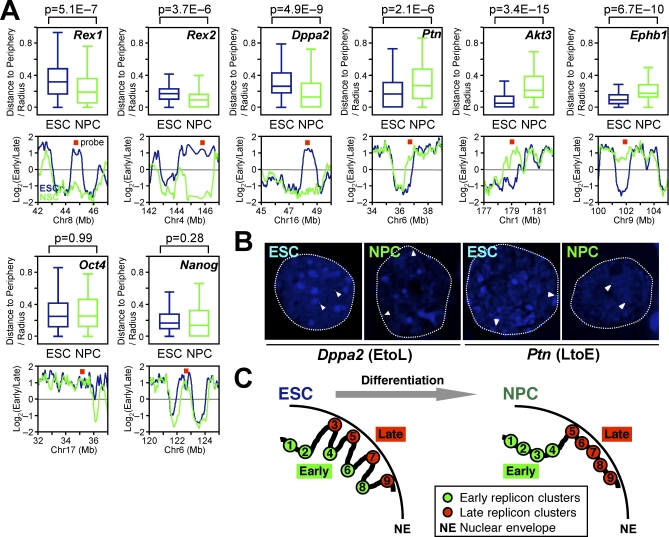
Figure 8. Global Temporal Reorganization of Replication Domains Reflects Spatial Reorganization in the Nucleus.
(A and B) Analysis of subnuclear positions of eight genomic regions by 3D-FISH in ESCs and NPCs. Box plots show radial distance to the nuclear periphery, where 0 and 1 represents the periphery and the center of the nucleus, respectively. Replication-timing profiles and the probe positions (red squares) are also displayed. Three EtoL domains (Rex1, Rex2, and Dppa2 domains) and three LtoE domains (Ptn, Akt3, and Ephb1 domains) move toward and away from the nuclear periphery, respectively, upon neural differentiation. Two EtoE domains (Oct4 and Nanog) do not change subnuclear positioning. Comparable results were obtained from two to four biological replicates, and the sum of all experiments is shown. A total of 90–234 alleles were measured per state.
(B) Representative FISH photographs (Dppa2 and Ptn). Dotted lines represent the rim of nuclear DAPI signals. Arrowheads represent DNA-FISH signals.
(C) A model for higher-order chromosomal organization in the nucleus during neural differentiation (see Discussion). Consider an example of two adjacent isochores, one GC rich (replicon clusters 1–4) and one AT rich (replicon clusters 5–9). In ESCs, replicon clusters with an unusual combination of GC content and gene density (clusters 3, 6, and 8) replicate differently and may be spatially separated from the rest of the isochore. During differentiation, there is an increased influence of isochore sequence features on replication timing, resulting in the temporal consolidation of these deviant domains and alignment of their replication timing to isochores, possibly accompanied by spatial reorganization.