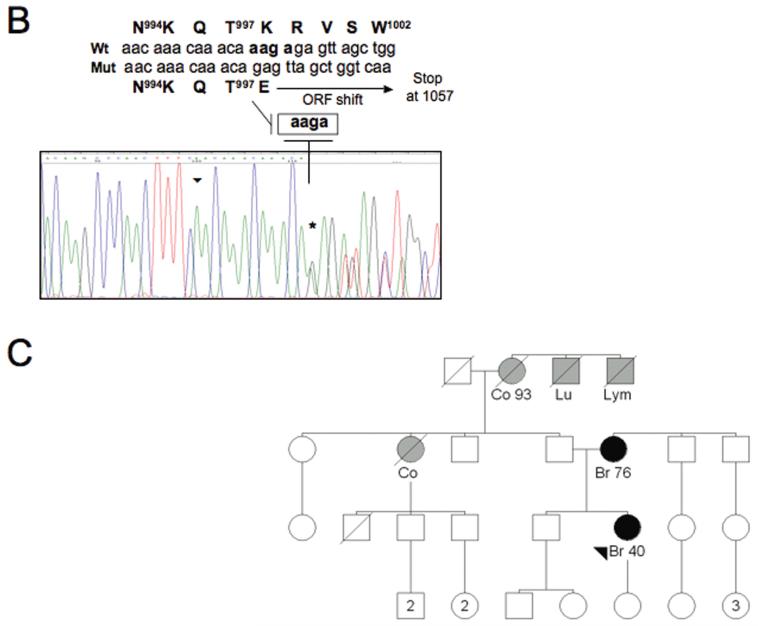
Figure 1. A novel BRIP1 sequence alteration identified in the family Pisa#88.
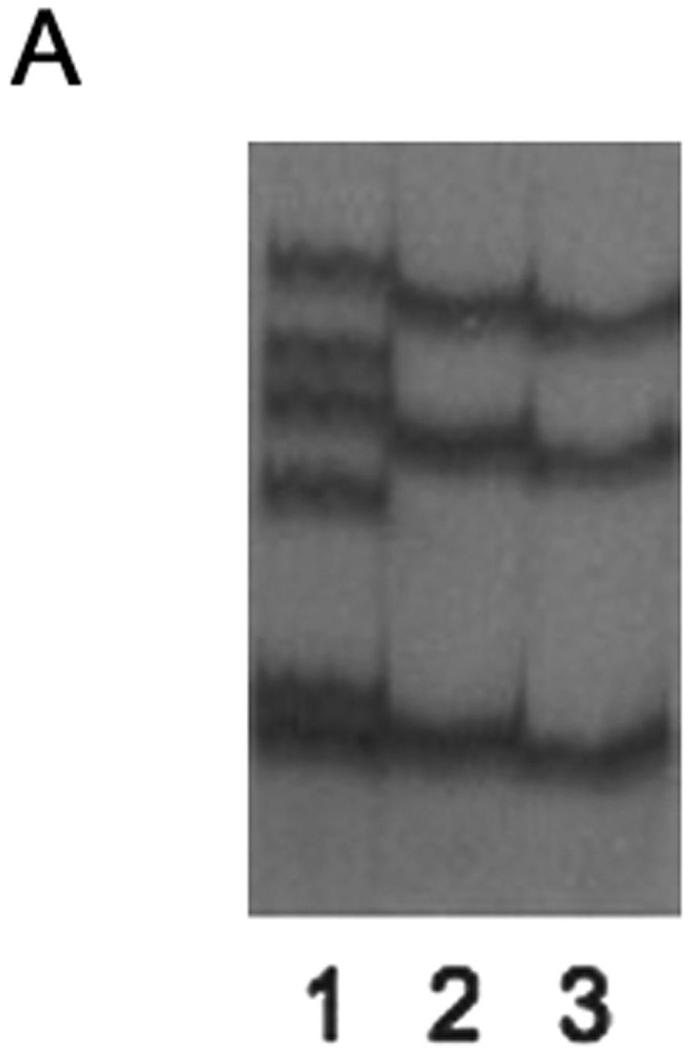
(A) Identification of a novel BRIP1 sequence variant. SSCP analysis after PCR amplification using primers located in BRIP1 exon 20 reveals an altered electrophoretic pattern in the sample corresponding to the proband of the family Pisa#88 (lane 1). Lanes 2 and 3, corresponding to the families Pisa#61 and Pisa#137, respectively, show a normal migration pattern, as ascertained in healthy control individuals (data not shown). (B) Characterization by sequence analysis of the BRIP1 gene alteration detected in the index case of the family Pisa#88. The chromatogram displays a 4 nucleotide-deletion, which starts at position 2992 (Genebank database sequence reference NM_032043, nucleotide numbering starting at codon 1), as indicated by an asterisk, and causes a shift of the open reading frame (ORF). The sequence of the wt and mutated alleles appear overlapped. The wt and mutated cDNA sequences are detailed (starting point marked by an arrowhead in the chromatogram). The corresponding amino acid sequences are reported, as well. The ORF shift (starting at codon 998) caused by the deletion results in a premature stop codon at position 1057. (C) Pedigree of the family Pisa#88, exhibiting the c.2992-2995delAAGA mutation. Black-filled symbols indicate individuals diagnosed with breast cancer (Br= breast cancer). Grey-filled symbols indicate individuals diagnosed with cancer other than breast (Co= colon cancer, Lu= lung cancer, Lym= lymphoma). The arrowhead indicates the index case.