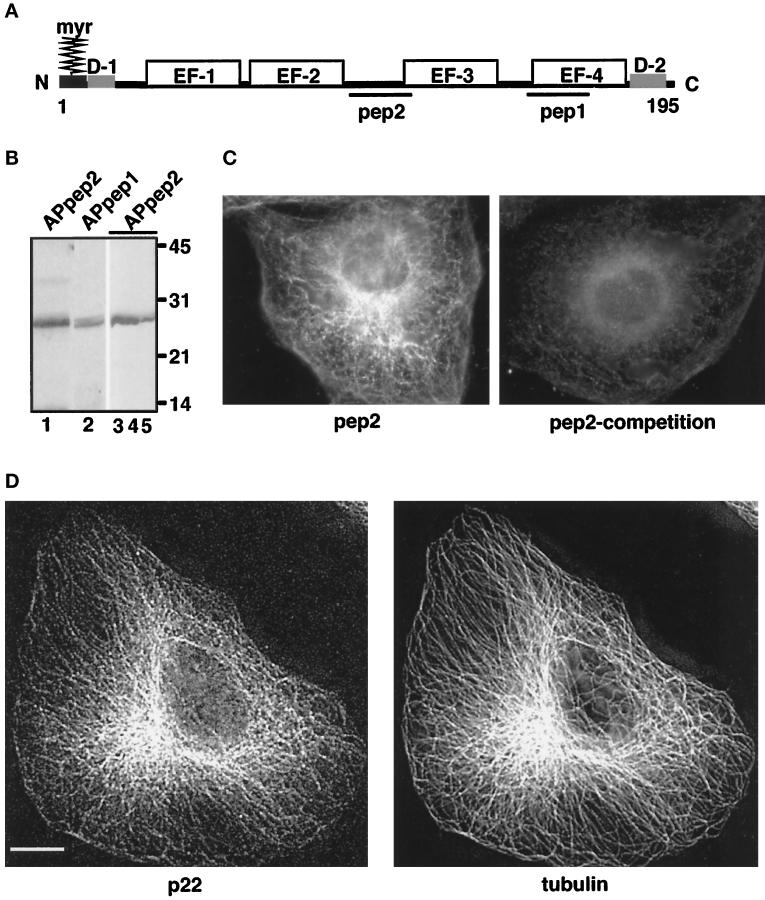
Figure 1.
Characterization of anti-p22 antibodies. (A) p22 conserved domains: myr, N-myristoylation consensus sequence; D-1, N-terminal acidic domain (9-LRDEELEEI-17); D-2, C-terminal domain (184-DVEQKMSIRFLH-195). EF-1, EF-2, EF-3, and EF-4 represent the four EF-hands of p22; pep1 and pep2 represent the sequences used to generate anti-p22 antibodies, APpep1 and APpep2, respectively. The D1 and D2 domains have 90% identity between the rat, human, and C. elegans sequences. (B) APpep2 antibodies recognize recombinant and rat liver p22. Rat liver cytosol (lanes 1 and 2), p22-rec (lane 3), p22-myr (lane 4), and p22-E134A (lane 5) were processed for immunoblotting using APpep2 (lanes 1, 3–5) or APpep1 antibodies (lane 2). A chemiluminescence image is shown. The molecular masses (kDa) are on the right. Note the consistent but retarded gel migration of p22 in relation to its predicted molecular mass (22 kDa). The same phenomenon is observed for rat liver p22 as well as for recombinant p22 and other EF-hand proteins such as recoverin. (C and D) p22 displays a microtubular pattern.NRK cells were processed for immunofluorescence using APpep2 antibodies (pep2) and anti-tubulin antibodies (tubulin). APpep2 antibodies incubated with pep2, before immunofluorescence, show a marked reduction in p22 staining (pep2-competition). Strong colocalization between p22 (p22) and microtubule (tubulin) staining patterns is detected in D. Images were collected by an Integrating CCD camera (PXL, 25°C cooled, Photometrics) in an IX-70 inverted Olympus fluorescence microscope using a 60 or 100× oil objective (W. M. Keck Center for Cellular Imaging, University of Virginia, Charlottesville, VA). Image processing was performed using Inovision ISee software Version 3.8 and Adobe Photoshop. Digital deconvolution was used to obtain images shown in D. Digital deconvolution was applied to 30 optical sections collected using DeltaVision Version 2.10 and a step of 0.4 μm. A representative optical section is shown. Bar, 10 μm.