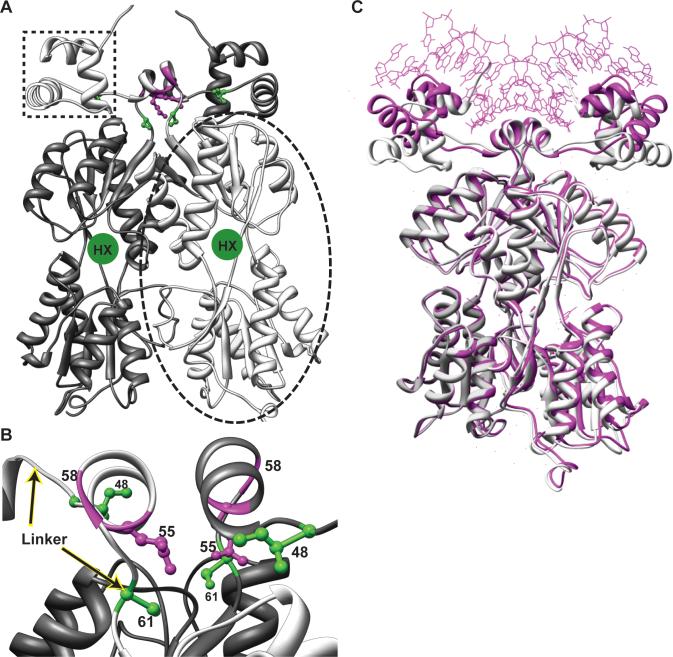
Figure 1. Model of the LLhP homodimer and positions of specificity determinants.
(A) The model of apo-LLhP was constructed as described in Materials and Methods and rendered with Chimera (50). One monomer is colored light gray and the other is dark gray. One DNA-binding domain is indicated with the dotted box; the two DNA-binding domains of a dimer are required for binding operator DNA. One regulatory domain is indicated within the dashed oval. The binding site for co-repressor hypoxanthine is indicated with a green circle in a cleft between the N- and C-subdomains (which are respectively at the top and bottom of the regulatory domain.) (B) The linkers of the LLhP dimer are shown, with the beginning and end indicated with arrows. The orientation is rotated ∼90° relative to (A). The central helical region of the linker is often called the “hinge helix”. The positions of linker specificity determinants for wild-type sites I48, Q55, and S61 are shown in ball-and-stick representation in green and magenta. Gly58 is indicated with a magenta ribbon for the backbone. None of the linker specificity determinants directly contact DNA in either the LacI or PurR structures. (C) The model for apo-LLhP (white) was aligned with the magenta structure of co-repressed PurR bound to DNA (1wet; (28)) using Chimera (50). Co-repressor is not shown in this figure. Note that the DNA-binding domains of apo-LLhP have moved relative to those of the DNA-bound PurR. Intriguingly, molecular dynamics simulations of the LacI DNA-binding domain show similar motions (8). Flexibility in the region spanning amino acids 45−50 is required for this motion, suggesting a possible mechanism for altering DNA-binding affinity by polymorphisms at position 48.