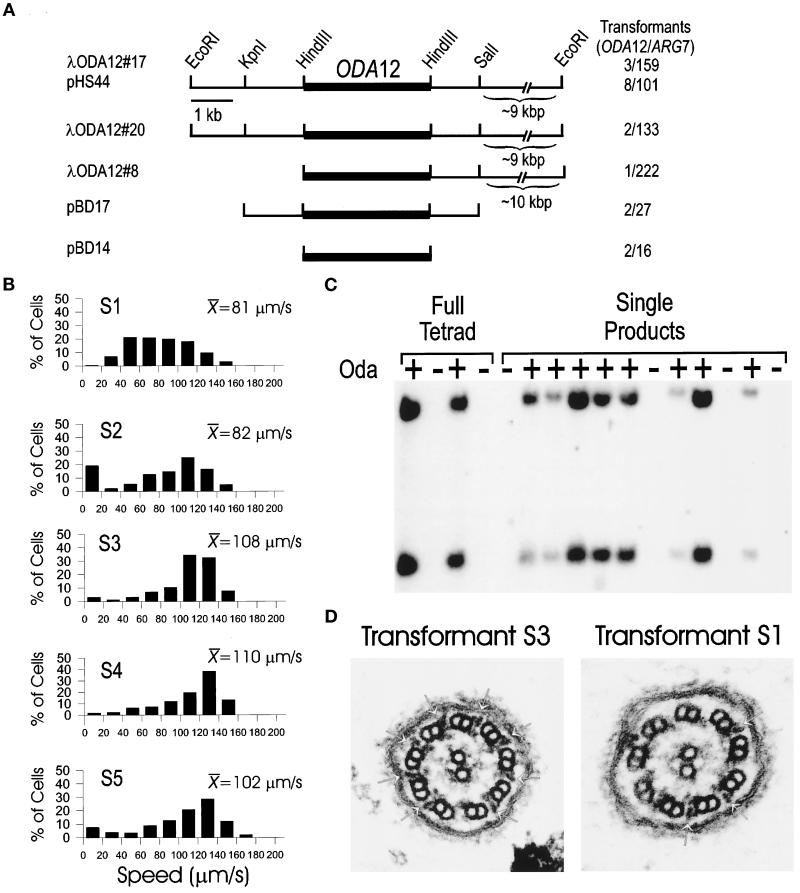
Figure 6.
Rescue of oda12 by transformation. (A) Map of the ODA12 locus. Three genomic λ phage clones (λODA12#8, λODA12#17, and λODA12#20) were isolated and mapped. The EcoRI sites at the ends of the clones are derived from the vector. The LC2 coding region (dark bar) was determined by hybridization with the cDNA clone. The 16-kbp insert from λODA12#17 was cloned into pKS+ and named pHS44. The 5.7-kbp KpnI–SalI and 3.1-kbp HindIII fragments of pHS44 were subcloned and named pBD17 and pBD14. The number of cell lines in which the Oda− phenotype was rescued out of the total number of ARG7 transformants screened is shown on the right. (B) Swimming speeds of transformed cell lines. The Speed Operator of the ExpertVision Motion Analysis system was used to calculate the swimming speed of five transformed lines (S1–S5). The histograms show the distribution of speeds within each sample. These are plotted as a percentage ofcells in each of 10 20-μm/s bins between 0 and 200 μm/s. The mean swimming speed (micrometers per second) for each sample also is shown. More than 100 cells were analyzed for each cell line. (C) Segregation of the exogenous copy of ODA12. oda12-1, arg7 strains were transformed with genomic clones of the ODA12 gene. One transformant (S20) that showed wild-type swimming speeds was mated to an oda12-1 strain of the opposite mating type, and tetrads were dissected. The offspring were scored for motility, and DNA was isolated from them. The DNA was cut with PvuII and analyzed by Southern blotting using the LC2 cDNA as a probe. The probe hybridizes with two bands in DNA from wild-type cells but does not hybridize with any band in DNA from oda12-1 cells (see Figure 1). Analysis of one full tetrad and a single product from each of 12 additional tetrads showed that fast-swimming cells (Oda+) had the LC2-hybridizing bands, whereas the slow-swimming cells (Oda−) were missing these bands. (D) Electron microscopy of flagella of transformed cells. Transformant S3, which swims at a speed similar to that of wild-type (cf. Figure 4 and B), has a full complement of outer dynein arms (left panel, arrows). In contrast, cells of transformed strain S1, which have a wide range of swimming speeds (Figure 6B), have variable numbers of outer dynein arms (right panel, arrows).