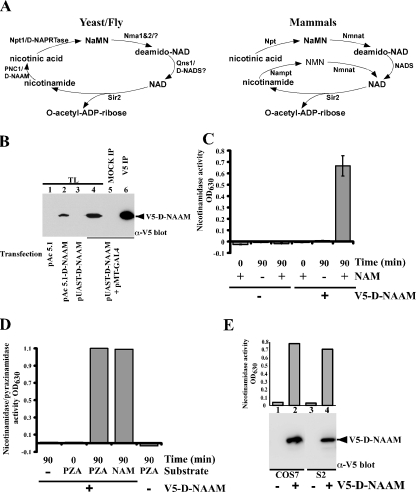
FIGURE 1.
The Drosophila homolog of yeast PNC1 encodes an active nicotinamidase. A, comparison of the NAD+ salvage pathway between mammals and yeast/flies. The nomenclature used is as in Rongvaux et al. (9) and Revollo et al. (8): PNC1, nicotinamidase; Npt1, nicotinic acid phosphoribosyltransferase; Nma1&2, nicotinic acid mononucleotide adenylyltransferase 1 and 2; Qns1, NAD synthetase; Nmnat, nicotinic acid mononucleotide adenylyltransferase; NADS, NAD synthetase; NaMN, nicotinic acid mononucleotide; NMN, nicotinamide mononucleotide. D-NAAM is our designation for Drosophila nicotinamidase. B, Drosophila S2 cells were transfected with carboxyl-terminally tagged V5-His-D-NAAM using the indicated expression vectors, and D-NAAM expression was determined in whole cell lysates (TL) or in V5 immunoprecipitates by V5 immunoblotting. C and D, V5 immunoprecipitates from S2 cells expressing a control vector or pAc 5.1-V5-His-D-NAAM were assayed for amidase activity using nicotinamide (NAM, C) or pyrazinamide (PZA, D) as a substrate. The activity is provided in A630 units corresponding to released ammonia. E, Drosophila S2 and mammalian COS-7 cells were transfected with control or pAc 5.1-V5-His-D-NAAM (S2 cells) or pExchange 5A-V5-His-D-NAAM (COS-7 cells) and nicotinamidase activity in V5 immunoprecipitates was analyzed as in C (top panel). D-NAAM recovery was determined using V5 immunoblotting (bottom panel).