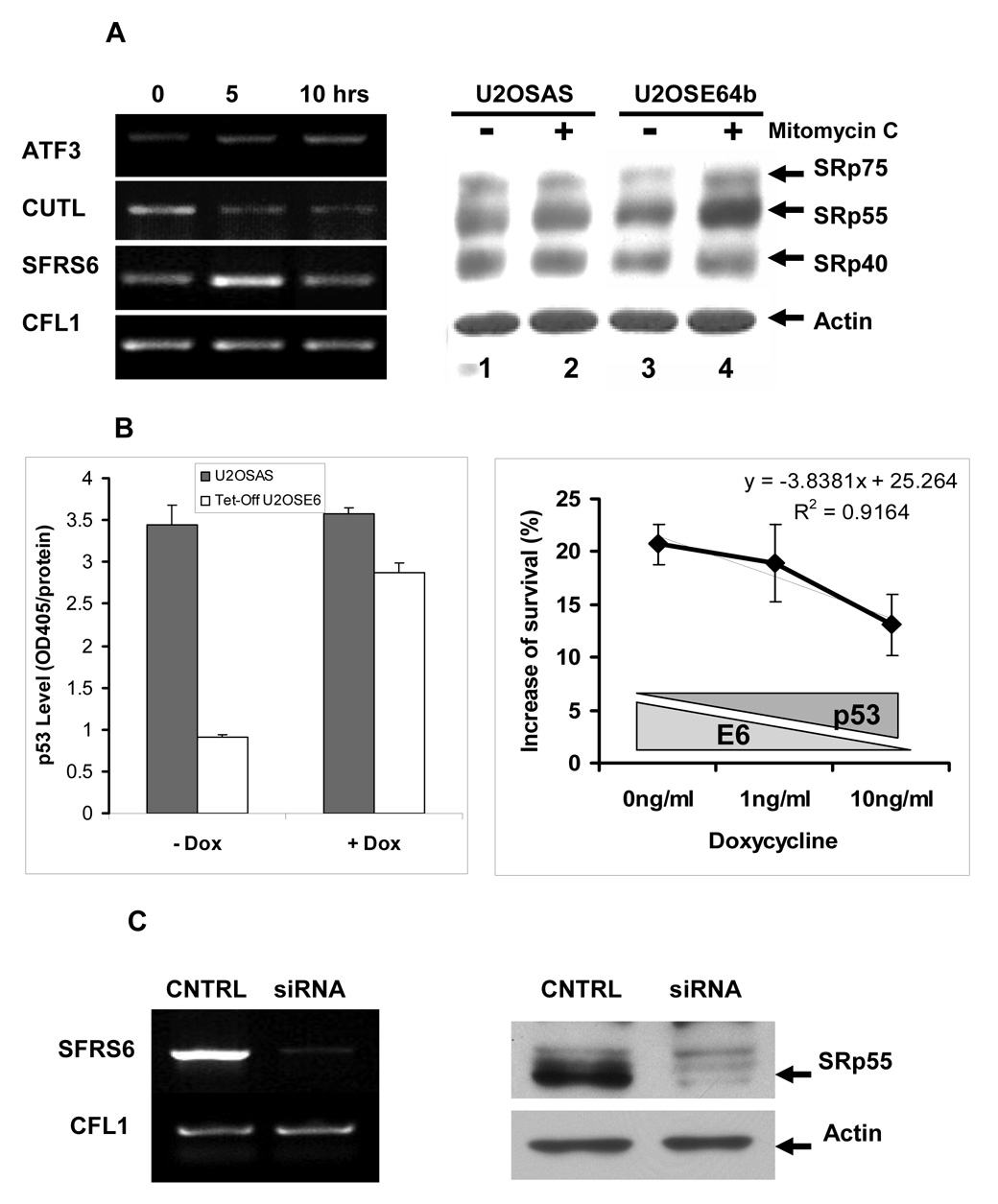
Figure 1. Characterization of RNA samples subjected to SpliceArray™ analysis and effect of SRp55 silencing on cell survival.
A. Left panel: Level of ATF3, CUTL, SFRS6, and CFL1 transcripts in U2OSE64b cells treated with 2 µg/ml mitomycin C for 5 and 10 hrs as determined by RT-PCR analysis. Right panel: The SRp55 protein encoded by the SFRS6 gene has elevated expression levels in U2OSE64b cells after incubation with mitomycin C (2 µg/ml) for 5 hrs, but not in the control U2OSAS cells (right panel). Lanes 1 and 3 show lysates of untreated U2OSAS and U2OSE64b cells, and lanes 2 and 4 show lysates of U2OSAS and U2OSE64b cells treated with 2 µg/ml mitomycin C for 5 hrs, respectively. The levels of splicing factors were determined by immunoblotting with monoclonal antibody 1H4 (ATCC, Manassas, VA). β-Actin antibodies were used for re-blotting to verify uniformity of sample loading. B. Left panel: Depletion of doxycycline (−Dox) in the medium effectively decreases p53 levels in the Tet-Off U2OSE6tet24 cell line but not in control U2OSAS cells. U2OSAS or Tet-Off U2OSE6 cells, grown in the presence or absence of 10 ng/ml Dox, were lysed and the lysates analyzed for p53 by ELISA. Right panel: Sensitivity of SRp55-depleted cells to mitomycin C treatment depends on the level of p53. U2OStetE624 cells, maintained in the presence of three concentrations of doxycycline, were transfected with SRp55 or scrambled siRNAs, then subjected to DNA damage by treatment with 4 µg/ml mitomycin C for 24 hrs. Viable cells were quantified using the MTT assay, and the differences between SRp55 silenced and control cells were compared by subtracting the number of viable cells in control scrambled siRNA samples from the number of surviving cells transfected with SRp55 siRNA. Means from triplicate measurements are shown, with the error bars representing the standard deviation. C. Left panel: Treatment of U2OS cells with SFRS6-specific siRNA effectively decreases the SFRS6 mRNA level. The level of SFRS6 mRNA was determined by RT-PCR and normalized against expression levels of the CFL1 gene. Right panel: Inhibition by siRNA leads to a decrease in the level of the SRp55 protein. Immunoblot of protein lysates isolated from control U2OS cells (CNTRL) and U2OS cells transfected with SFRS6 siRNA (siRNA). The β-actin antibodies were used to verify uniformity of sample loading.