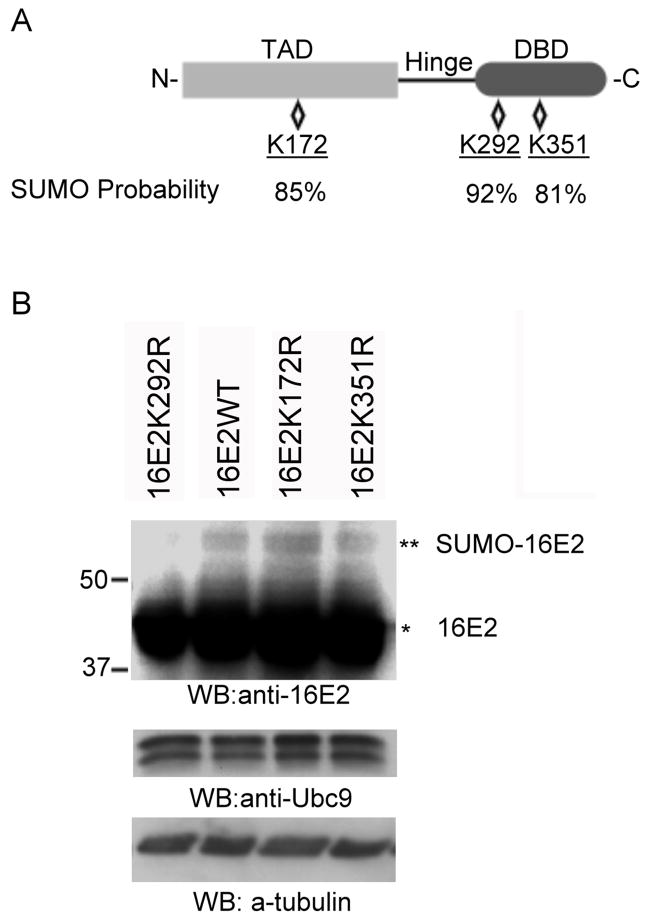
Figure 4.
Mapping of the sumoylation site in HPV16 E2. (A) Structure and predicted sumoylation sites in HPV16 E2. The relative locations of the N-terminal transactivation domain (TAD), the hinge region, and the C-terminal DNA-binding domain (DBD) are as shown. There are three lysine residues predicted to have a high probability for sumoylation with the probability of the sumoylation indicated below each residue. (B) HeLa cells were transfected with a combination of 1.5 μg of pcDNA3.1/HA-SUMO3 and 1.5 μg of pcDNA5/FRT/TO/HA-Ubc9 together with 3 μg of pWEB-16E2 wildtype (16E2WT, left panel), pWEB-16E2K292R (16E2K292R, left panel), pWEB-16E2K172R (16E2K172R, left panel), pWEB-16E2K351R (16E2K35 1R, left panel), or pWEB-16E2K292RK172R (16E2K292RK172R, right panel). Cells were processed as in Figure 3 and were analyzed by Western blotting with anti-16E2, anti-Ubc9 antibodies, or anti-tubulin (loading control) as indicated. Ubc9 served as the control for comparing transfection and expression efficiency. The single asterisk (*) indicates the unmodified form of either HPV16 E2, and the double asterisk (**) indicates the sumoylated form.