Figure 1.
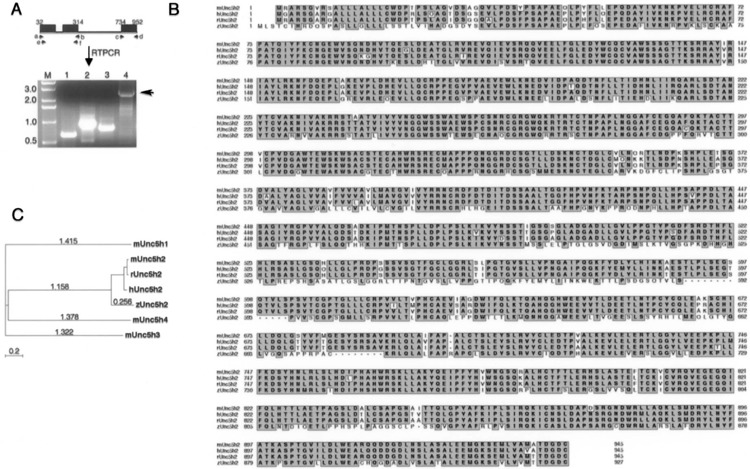
Cloning and phylogenetic analysis of unc5b.(A) The black boxes depicts the three exons that correspond to human unc5b amino acids, which hit putative zebrafish nucleotide sequence in Tblastn searches of Sanger zebrafish genome database. The six primer pairs (a to f) used for PCR are shown by a small black arrow. Lane M: marker (kb); lane 1: c and d; lane 2: a and b; lane 3: e and f: lane 4: a and d. Arrow indicates the 2.8-kb fragment of unc5b, which encompasses most of the unc5b gene except the first 35 amino acids. (B) Phylogenetic comparisons of zebrafish unc5b to unc5b members in other species and to mouse unc5 family (1, 3, and 4) are shown. The CLUSTALW program, as part of the MacVector 7.0 Software package was used for generating the tree and the accession numbers for sequences used here are zUnc5b (DQ365815), mUnc5h1 (NP_694771.1), mUnc5h3 (NP_033498), mUnc5H4 (NP_694775), mUnc5b (NP_084046), rUnc5b (NP_071543), and hUnc5b (NP_734465). Evolutionary tree was built using the “Neighbor joining” method, which does not assume constant divergent rates among sequences, and evolutionary distances were calculated using “Best tree” method, which estimates the number of substitutions per site under the assumption that the distribution follows a poisson distribution. (C) Sequence homology comparisons with different unc5b family members are shown. The human, mouse, and zebrafish unc5b amino acid sequences were aligned using CLUSTALW program in MacVector 7.0. Pairwise comparisons between human and mouse sequences showed 90% homology while between human or mouse to zebrafish unc5b was 65%. Unc5h1, Unc5h2, Unc5h3, and Unc5h4 is the old nomenclature for Unc5a, Unc5b, Unc5c, and Unc5d, respectively.
