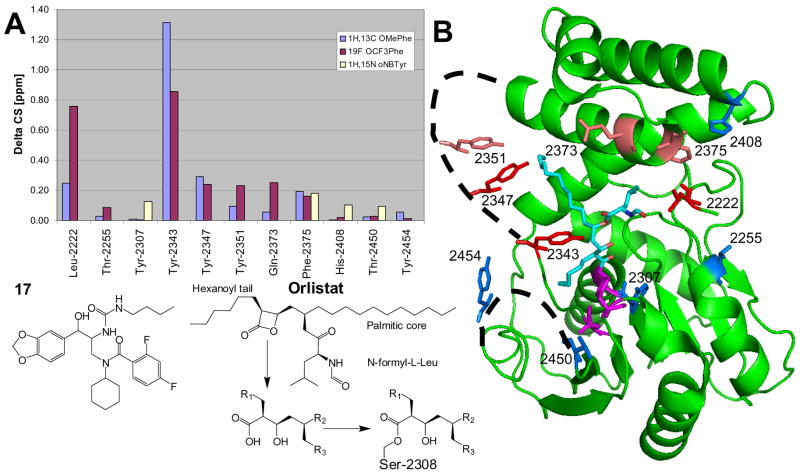
Figure 8.
Chemical shift mapping of tool compound 17 binding to FAS-TE. A) Combined chemical shift data. Chemicals shift changes ΔCS of 19F resonances (OCF3Phe; purple) were scaled relative to 1H by the gyromagnetic ratio while 1H-13C (OMePhe; violet) and 1H-15N (oNBTyr; pale yellow) chemical shifts were combined using the expression ΔCS = [(Δ1H)2 + (0.252*Δ13C)2]0.5 and ΔCS = [(Δ 1H)2 + (0.102* Δ 15N)2]0.5, respectively. OMePhe and OCF3Phe at Tyr-2307 block binding (see text). For oNBTyr mutants only a limited set of chemical shift changes could be compiled because of conformational exchange (see text). B) Structure of the covalent orlistat complex (2PX6.pdb).30 The average chemical shift changes induced by the binding of 17 are calculated for each unnatural amino acid and color-coded for each position: Δ CS < 0.1 ppm, blue; 0.1 to 0.2 ppm, salmon; > 0.2 ppm, red. Disordered loops are indicated by dashed lines. The active site residues Ser-2308, Asp-2338 and His-2481 are shown in magenta.