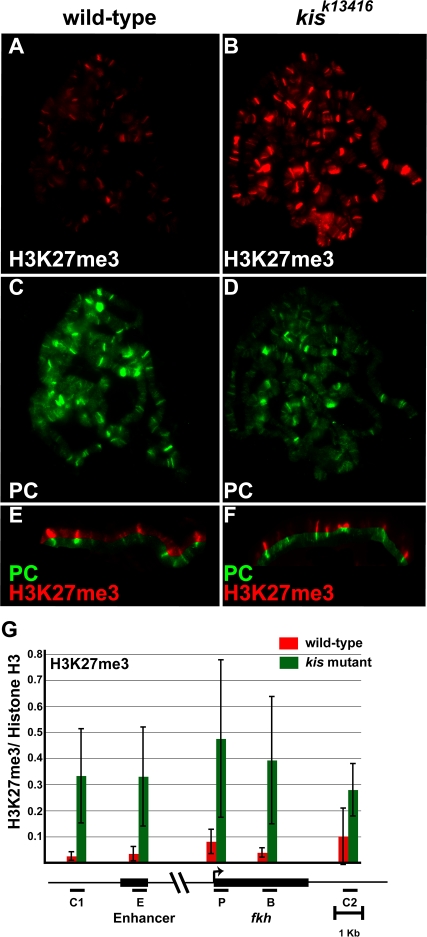
Figure 9. Mutations in kis lead to increased H3K27 methylation.
A–D) The level and distribution of H3K27me3 (A, B, red) and PC (C, D, green) on salivary gland polytene chromosomes of wild-type (A, C) and kisk13416 (B, D) larvae was examined by double-label indirect immunofluorescence microscopy. Split images of the distributions of H3K27me3 (red) and PC (green) on wild-type (E) and kisk13416 (F) polytene chromosomes are shown. Loss of kis function results in an increase in the levels of H3K27me3 without altering the distribution of this modification on polytene chromosomes. For panels E and F, the levels of H3K27me3 were independently processed using Adobe Photoshop software to facilitate the comparison of the methyl mark and PC. G) The distribution of H3K27me3 over the fkh gene was determined by ChIP using chromatin isolated from the salivary glands of wild-type (red bars) or kisk13416 (green bars) larvae. A map of the fkh gene is shown below the X axis; black bars represent the primers used to amplify the following regions: C1: region upstream of fkh, E: fkh enhancer, P: fkh transcription start site, B: fkh body, C2: region downstream of fkh. The ratio of DNA immunoprecipitated with antibodies against H3K27me3 and histone H3 are shown for each region. The bars represent the average of three independent biological experiments with the corresponding standard deviations. The loss of KIS-L function leads to a significant increase in H3K27me3 over the entire fkh region.