Abstract
Transcriptional dysregulation has emerged as a core pathologic feature of Huntington's disease (HD), one of several triplet-repeat disorders characterized by movement deficits and cognitive dysfunction. Although the mechanisms contributing to the gene expression deficits remain unknown, therapeutic strategies have aimed to improve transcriptional output via modulation of chromatin structure. Recent studies have demonstrated therapeutic effects of commercially available histone deacetylase (HDAC) inhibitors in several HD models; however, the therapeutic value of these compounds is limited by their toxic effects. Here, beneficial effects of a novel pimelic diphenylamide HDAC inhibitor, HDACi 4b, in an HD mouse model are reported. Chronic oral administration of HDACi 4b, beginning after the onset of motor deficits, significantly improved motor performance, overall appearance, and body weight of symptomatic R6/2300Q transgenic mice. These effects were associated with significant attenuation of gross brain-size decline and striatal atrophy. Microarray studies revealed that HDACi 4b treatment ameliorated, in part, alterations in gene expression caused by the presence of mutant huntingtin protein in the striatum, cortex, and cerebellum of R6/2300Q transgenic mice. For selected genes, HDACi 4b treatment reversed histone H3 hypoacetylation observed in the presence of mutant huntingtin, in association with correction of mRNA expression levels. These findings suggest that HDACi 4b, and possibly related HDAC inhibitors, may offer clinical benefit for HD patients and provide a novel set of potential biomarkers for clinical assessment.
Keywords: chromatin, neurodegenerative, epigenetic, therapeutic, trinucleotide
Huntington's disease (HD) is an autosomal-dominant neurologic disorder caused by a CAG repeat expansion within the coding region of the HD gene (Htt), resulting in a mutant protein (htt) with a lengthened polyglutamine tract (1). Mutant htt protein has been shown to disrupt transcription by multiple mechanisms, but it is unclear which are most important to pathology (2–4). By interacting with specific transcription factors, htt can alter the expression of clusters of genes controlled by those factors. For example, several genes driven by Sp1, which has been shown to interact with htt (5, 6), show decreased mRNA expression in human HD and in mouse models of HD (7). Alternatively, htt may have more global effects on transcription by disrupting core transcriptional machinery (8, 9) or by altering posttranslational modifications of histones, resulting in condensed chromatin structure (10–13). Understanding the basis for transcriptional dysregulation is important for choosing appropriate drug-discovery strategies.
Manifestations of transcriptional dysregulation are evident from several gene-profiling studies, which have revealed alterations in the expression of large numbers of genes in the brains of different HD mouse models and in human subjects with HD (7, 14–16). Many of the expression changes in mouse models are observed in early stages of illness before the onset of symptoms, suggesting that gene expression alterations may be pathogenic. Because of the extent of gene expression alterations in HD, most of which are decreases in expression, agents that improve transcriptional activity on a broad scale may represent an important therapeutic approach for HD. In addition, the evidence for chromatin-based transcriptional repression in HD suggests that inhibitors of histone deacetylase (HDAC) enzymes, which act in concert with histone acetyltransferase enzymes to modulate gene transcription, may represent useful treatments for HD.
Previous studies have examined the potential therapeutic effects of the HDAC inhibitors suberoylanilide hydroxamic acid (SAHA) (17), sodium butyrate (18), and phenylbutyrate (19) in HD mouse models. Despite showing promise in ameliorating the phenotype in different HD mouse models, the utilities of these compounds, as well as their analogues, are limited by toxicity. Toxicity studies of various HDAC inhibitors, including SAHA, have demonstrated widespread effects in human cancer cells in vitro, including activation of proapoptotic and inhibition of antiapoptotic pathways, stimulation of cell differentiation, and induction of growth arrest (20–22). These features have led to the approval of SAHA for use in human cancer clinical trials (22); however, such properties might be expected to exacerbate symptoms in neurodegenerative disorders, such as HD.
We have developed a class of benzamide-type HDAC inhibitors that show promising results in Friedreich's ataxia disease models (23, 24). These compounds are structurally related to the well-known HDAC inhibitor SAHA but are not hydroxamic acids and, unlike SAHA, were found to increase expression of the frataxin gene in lymphocytes from Friedreich's ataxia patients (23). From a panel of these novel HDAC inhibitors, we have further characterized the therapeutic potential in HD mice for one selected compound, HDACi 4b. Our cell culture findings indicate that HDACi 4b shows a low toxicity profile, whereas our in vivo studies on R6/2 transgenic mice, which is the most widely used model for preclinical trials (25, 26), demonstrate therapeutic efficacy in preventing motor deficits and neurodegenerative processes. We further report that HDACi 4b treatment ameliorates gene expression abnormalities detected by microarray analysis in these mice.
Results
In Vitro Toxicity Profile of HDACi 4b.
We evaluated the cytotoxic effects of HDACi 4b treatment on cell cycle parameters in human lymphoblast cell cultures. Cells were treated with increasing concentrations of HDACi 4b (1–125 μM) for 72 h and then assessed by FACS analysis of propidium iodide-stained nuclei. This analysis demonstrated no cell-cidal effects at concentrations <50 μM and only cell-static effects at concentrations >20 μM [supporting information (SI) Fig. S1]. No apoptotic effects of HDACi 4b were observed, except at concentrations >50 μM (Fig. S1), which are 10-fold higher than that previously reported for SAHA using similar cell types and methodologies (27). Importantly, at the highest concentration of 0.125 mM HDACi 4b, only 14% of the total cells gated were observed to be apoptotic (Fig. S1). Given an IC50 value of ≈1 μM for HDACi 4b-mediated inhibition of HDAC activity (as measured in HeLa cell nuclear extracts), the concentrations imparting toxic effects are 20–50-fold higher.
HDACi 4b Improves Disease Phenotype in R6/2300Q Transgenic Mice.
We first verified that HDACi 4b can modify the histone acetylation status in the CNS in vivo. A single injection of HDACi 4b (100 mg/kg, s.c) in normal mice resulted in a nearly twofold increase in CNS histone H4 acetylation (Fig. S2). We next developed a formulation for improving water solubility of HDACi 4b by complexing with 2-hydroxypropyl-β-cyclodextrin (HOP-β-CD), similar to that reported previously (17); this allowed for chronic dosing of HDACi 4b in drinking water, corresponding to a dose of 150 mg/kg/day. R6/2300Q transgenic mice and WT littermate controls were treated with HDACi 4b for 67 days beginning at 4 months of age, which is after the onset of motor dysfunction, as evidenced by rotarod deficits at baseline training (Fig. 1G). The effect of HDACi 4b administration on the R6/2300Q phenotype was assessed by several measurements, including motor function, physical appearance, and body weight.
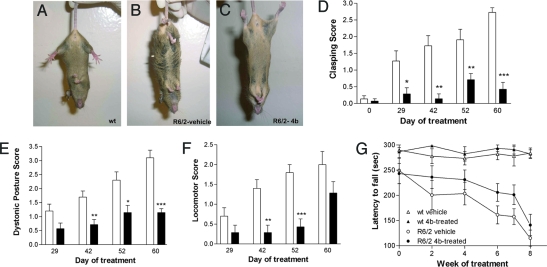
Fig. 1.
Behavioral phenotypes of vehicle- and HDACi 4b-treated mice. Hindlimb clasping phenotype of WT mice (A) and R6/2300Q transgenic mice treated with vehicle (B) or HDACi 4b (C). The differences between HDACi 4b-treated (filled bars) and vehicle-treated (open bars) R6/2300Q transgenic mice are shown for the clasping phenotype (D), dystonic posture (E), and locomotor activity (F) over the time of drug treatment. Two-way ANOVAs were used to determine the effect of HDACi 4b treatment and treatment duration on each of these measurements. *, P < 0.05; **, P < 0.001; ***, P < 0.0001. (G) Rotarod performance of HDACi 4b-treated and vehicle-treated WT and R6/2300Q transgenic mice throughout the treatment duration. Time-point 0 reflects baseline performance. Two-way ANOVA revealed significant differences between WT and R6/2300Q transgenic mice with time (P < 0.0001), as well as a significant effect of drug treatment in R6/2300Q transgenic mice (P < 0.05). Bars represent mean score ± SEM (n = 7 to 8 per group).
Motor function abnormalities were determined by measuring clasping phenotype, general locomotion, and rotarod performance. As expected, R6/2300Q transgenic mice exhibited significant deficits in motor behavior with increasing treatment duration (i.e., age) (Fig. 1). However, significant prevention or amelioration of these deficits were observed with HDACi 4b treatment for the hindlimb clasping test [F(1,80) = 64.07; P < 0.0001], generalized locomotor behavior [F(1,60) = 26.81; P < 0.0001], and rotarod performance [F(1,59) = 4.77; P = 0.032)] (Fig. 1). Most notably, as shown in Fig. 1A, HDACi 4b-treated mice show almost no clasping phenotype. We also assessed the effects of HDACi 4b on physical appearance, which can be quantified, in one sense, by monitoring changes in dystonic posture exhibited by R6/2300Q transgenic mice. We found that HDACi 4b treatment significantly prevented the development of kyphosis in R6/2300Q transgenic mice [two-way ANOVA: F(1,60) = 43.76; P < 0.0001] (Fig. 1E). We further found that HDACi 4b treatment resulted in improved coat appearance, gait stability, and overall motility in R6/2300Q transgenic mice (Movie S1, Movie S2, and Movie S3).
The effects of HDACi 4b treatment on body weight on each genotype were determined by using two-way ANOVA. All R6/2300Q transgenic mice showed a decrease in body weight with duration of treatment (i.e., age) [F(22,228) = 9.63; P < 0.0001]. However, when comparing the two groups of transgenic mice, we found a significant effect of HDACi 4b treatment to attenuate body-weight decline [F(1,228) = 48.36; P < 0.0001] (Fig. 2A). HDACi 4b also had a small but significant effect on body weight in WT mice [F(1,256) = 9.24; P < 0.002] (Fig. 2B).
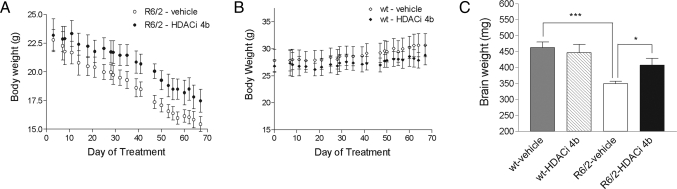
Fig. 2.
Body weights over time of R6/2300Q transgenic (A) and WT (B) mice throughout HDACi 4b treatment. Symbols represent mean body weight ± SEM in vehicle-treated mice (open symbols) and HDACi 4b–treated mice (filled symbols). R6/2300Q transgenic mice treated with HDACi 4b weighed significantly more than vehicle-treated R6/2300Q mice as determined by two-way ANOVA (P < 0.0001). (C) Effect of HDACi 4b treatment on overall brain weights of WT and R6/2300Q transgenic mice. Significant differences were determined by one-way ANOVA and Student's t test. ***, P < 0.001; *, P < 0.05.
HDACi 4b Shows Neuroprotective Effects.
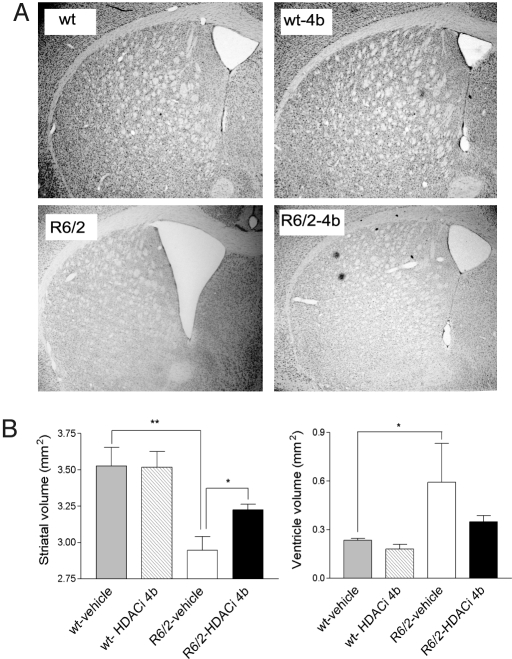
At the end of drug treatment, which was close to the lifespan of the mice (6 months of age), neuroprotective effects of HDACi 4b treatment were assessed. Overall, there was a 22.9% reduction in brain weight of vehicle-treated R6/2300Q mice compared with WT littermates at age 6 months (356.4 ± 10.19 mg vs. 462 ± 18.32 mg for R6/2300Q and WT mice, respectively, P = 0.003) (Fig. 2). However, brains from HDACi 4b-treated mice weighed significantly more when compared with those from vehicle-treated R6/2300Q mice (407.3 ± 21.74 mg vs. 356.4 ± 10.19 mg for drug-treated and vehicle-treated R6/2300Q mice, respectively, P = 0.045) (Fig. 2C). Furthermore, HDACi 4b treatment ameliorated the gross striatal atrophy and ventricular enlargement that was observed in the vehicle-treated R6/2300Q mice (Fig. 3). Remarkably, brains from HDACi 4b-treated mice were indistinguishable from those from vehicle- or HDACi 4b-treated WT mice (Fig. 3). No apparent difference in the number of huntingtin-positive striatal nuclear aggregates, as determined by immunohistochemistry using the EM48 anti-huntingtin antibody, was observed between vehicle- and drug-treated R6/2300Q mice (data not shown), consistent with previous studies using sodium butyrate, phenylbutyrate, or SAHA (17–19).
Fig. 3.
Effects of HDACi 4b on brain gross morphology. Volumetric analysis of striatum and lateral ventricles was performed on mice at 6 months of age. (A) Representative images of brain at ≈0.62 bregma for the indicated conditions. (B) Quantification of striatal area and lateral ventricles. Bar graphs represent mean ± SEM for WT mice and R6/2300Q mice (n = 6 per group). Significant differences were determined by one-way ANOVA and Student's t test. **, P < 0.001; *, P = 0.05. The effect of drug treatment on ventricular volume did not reach significance.
HDACi 4b Treatment Ameliorates Gene Expression Abnormalities in R6/2300Q Transgenic Mice.
To evaluate whether HDACi 4b treatment alters gene expression profiles in the CNS of WT and R6/2300Q transgenic mice, we performed microarray analysis on striatum, cortex, and cerebellum of 5-month-old vehicle- and HDACi 4b-treated mice. Hierarchal clustering of the samples based on interarray Pearson correlation showed sample clustering according to brain region and genotype, as expected (Fig. S3). Consistent with results from previous microarray studies (16), R6/2300Q transgenic mice showed a significant number of differentially expressed genes, with cortex and striatum showing an overrepresentation of down-regulated genes. The top expression differences observed in transgenic vs. WT mice at P < 0.005 (n = 988, 724, and 979 probes for striatum, cortex, and cerebellum, respectively) are listed in Dataset S1.
Because our R6/2 line carries a greater CAG repeat than the parent line, we assessed to what extent these mice recapitulate the gene expression profiles detected in human HD by comparing gene transcriptome profiles from the striatum of R6/2300Q transgenic mice with previously published microarray data from human HD caudate samples (15). Human orthologues of the top 250 differentially expressed mouse genes in the striatum (including both increases and decreases) were mapped and their expression differences screened in the human HD dataset provided by Hodges et al. (15). In our dataset, 77 of the top 142 down-regulated genes (54.2%) and 39 of the top 80 up-regulated genes (39%) showed concordance expression changes, defined as those showing a significant difference in the human dataset (P < 0.05) and the same direction of change in both mouse and human. A concordant coefficient was calculated, taking into account both concordant and discordant changes, as described previously (16) (SI Methods). The concordance coefficient for our dataset was 0.378 for down-regulated genes and 0.327 for all genes, which is consistent with previous studies reporting statistically significant concordance coefficients for other HD mouse models of 0.190 to 0.490 (with the related R6/1 and R6/2 lines equaling 0.350 and 0.405/0.490, respectively) (16). Hence, our findings indicate that R6/2300Q transgenic mice exhibit expression profiles highly correlated with those observed in human HD.
HDACi 4b treatment caused a less severe effect on overall gene expression, compared with the dramatic effect caused by the presence of the mutant Htt gene. Of the top disease-induced gene expression abnormalities (at P < 0.005), ≈20% showed a change toward the WT level after HDACi 4b administration (Fig. S4). To determine more inclusive lists of potentially therapeutically relevant expression changes, we identified the intersection between disease- and drug-induced expression changes at P < 0.05, which resulted in a list of 320, 167, and 137 genes for the cerebellum, striatum, and cortex, respectively. Depending on the region, 85%–94% of these genes showed a trend toward normalization, with one-third being completely normalized by HDACi 4b treatment (Fig. 4). These genes (provided in Dataset S2) could be considered as potential candidate biomarkers for treatment effectiveness. Interestingly, the expression of different sets of genes was altered by HDACi 4b treatment in WT compared with HD transgenic mice in each brain region, with only a <10% overlap of drug-induced differential gene expression between the genotypes (Fig. S5), indicating tissue-specific effects. HDACi 4b treatment did not have any effects on the expression of the endogenous Htt gene in R6/2300Q transgenic mice, nor the human HTT transgene, as determined by real-time PCR analysis (data not shown).
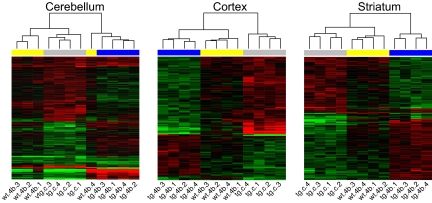
Fig. 4.
Heatmaps depicting the probes that are differentially expressed in R6/2300Q mice vs. WT (P < 0.05, gray bar) and in HDACi 4b-treated vs. untreated R6/2300Q transgenic mice (P < 0.05, blue bar). The HDACi 4b effect on WT mice in these genes is marked by the yellow bars. Log2-transformed ratios were subjected to two-dimensional hierarchical clustering. Each colored pixel represents an individual ratio from a single subject (four replicates per condition) compared with the average of controls. Relative decreases in gene expression are indicated by green and increases in expression by red. The vast majority of these genes show a trend toward normalization after HDACi 4b treatment. Complete normalization is reached for 100 (31%, cerebellum), 45 (33%, cortex), and 56 (33%, striatum) probes.
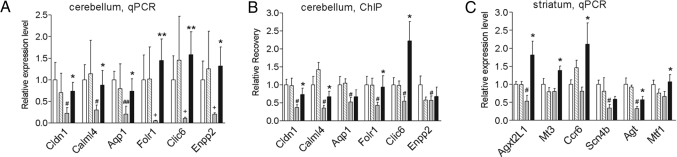
Expression differences for a diverse group of selected genes were validated by real-time PCR analysis. From the microarray data, the cerebellum showed the most robust treatment effect, which is interesting in light of studies suggesting that the cerebellum may play a more significant role in the symptomatology of HD than previously thought (28). Expression deficits for aquaporin 1 (Aqp1), claudin 1 (Cldn1), calmodulin-like 4 (Calml4), folate receptor 1 (Folr1), chloride intracellular channel 6 (Clic6), and ectonucleotide pyrophosphatase/phosphodiesterase 2 (Enpp2), detected in the cerebellum of 5-month-old R6/2300Q transgenic mice compared with age-matched WT mice, were almost completely reversed by HDACi 4b treatment (Fig. 5A), validating the array. We next assessed the histone acetylation status of these down-regulated genes in the same sets of mice by using ChIP analysis in combination with real-time PCR. Compared with WT mice, hypoacetylation of histone H3 was detected specifically in association with these six down-regulated genes in R6/2300Q transgenic mice (Fig. 5B). After treatment with HDACi 4b, significant increases in histone H3 acetylation was detected for Cldn1, Calml4, Folr1, and Clic6 (Fig. 5B). Significant effects of HDACi 4b treatment on the expression of alanine-glyoxylate aminotransferase 2-like 1 (Agxt2l1), metallothionein 3 (Mt3), chemokine receptor 6 (Ccr6), metal response element binding transcription factor 1 (Mtf1), and angiotensinogen (Agt) in the striatum of R6/2300Q mice were also validated by real-time PCR analysis (Fig. 5C).
Fig. 5.
HDAC inhibitor treatment reverses mRNA abnormalities and increases acetylated H3 in association with genes down-regulated in R6/2300Q transgenic mice. Real-time PCR analysis was performed for the indicated genes on RNA samples from the cerebellum (A) or striatum (C) of vehicle- (white bars) or HDACi 4b–treated (striped bars) WT mice and vehicle- (gray bars) or HDACi 4b–treated (filled bars) R6/2300Q transgenic mice. Data are depicted as fold change of the mean expression level ± SEM (n = 4 mice per group). The relative abundance of each gene expression was normalized by using Hprt. (B) Chromatin immunoprecipitation was performed with anti-acetylated histone H3 (AcH3) and anti-histone H3 antibodies on the cerebellum of vehicle- or HDACi 4b-treated WT and R6/2300Q transgenic mice (same key as in A and C). Real-time PCR was performed with primers for the promoter region of Cldn1 and Aqp1 and the transcribed region of Calml4, Folr1, Clic6, and Enpp2. Recovery was normalized for Gapdh, and data are shown as the ratio of the recovery for AcH3 and H3 and normalized for vehicle-treated WT mice ± SEM. Student's t tests were used to determine significant differences in gene expression or recovery levels. The effect of drug treatment on expression of the sodium channel, type IV-β gene (Scn4b) in the striatum of HD mice did not reach significance. # denotes significantly different values between R6/2300Q transgenic and WT mice at P < 0.05, with + denoting P < 0.08; * denotes significantly different values between HDACi 4b- and vehicle-treated R6/2300Q mice at P < 0.05 and ** at P < 0.01.
Discussion
In this study, we report beneficial effects of a novel benzamide-type HDAC inhibitor, HDACi 4b, on the disease phenotype of R6/2300Q transgenic mice at the behavioral, physical, pathologic, and molecular levels. Importantly, we find very low toxic effects of HDACi 4b associated with these outcomes or at therapeutic doses in vitro. HDACi 4b-treated mice showed significantly improved movement and coordination and delayed weight loss over vehicle-treated animals, despite initiation of treatment after the onset of motor deficits. This finding is in contrast to previous studies investigating the effects of phenylbutyrate on N171–82Q transgenic mice, in which beneficial effects on these parameters, including rotarod, were not found with treatment beginning after symptom onset (19). HDACi 4b treatment also dramatically improved the physical appearance of, and prevented dystonic posture exhibited by, R6/2300Q transgenic mice (see Movie S1, Movie S2, Movie S3, and Fig. 1). Such effects have not been reported in response to other HDAC inhibitors previously tested in HD mouse models (17–19). Overall, these findings suggest that HDACi 4b treatment may slow the progression of symptoms in humans, who are often diagnosed after symptom presentation.
At the molecular level, we find that HDACi 4b treatment results in both increases and decreases in gene expression of a limited number of genes, rather than having global effects on transcription. This effect is likely because of specificity of HDACi 4b for particular HDAC isoforms, of which at least 11 are expressed in the rodent brain (29). These findings are also consistent with previous studies indicating that commercially available HDAC inhibitors can have both positive and negative effects on gene expression (reviewed in ref. 22). Because HDAC inhibition is expected to result primarily in increased gene transcription, the down-regulation of certain genes may represent a secondary effect.
The therapeutic effects of HDACi 4b likely result from effects on histone acetylation and gene transcription of only a subset of genes that are abnormally expressed in disease. Previous studies have demonstrated hypoacetylation of histones specifically in association with the dopamine D2 receptor and enkephalin, which are down-regulated in R6/2 transgenic mice (30). The authors of these studies further demonstrated reversal of histone hypoacetylation and decreased expression of these genes after treatment with the HDAC inhibitor phenylbutyrate (30). In the present study, we show similar effects for six genes in response to HDACi 4b treatment. In addition, we find complete reversal of the htt-induced transcription aberrations for subsets of genes in all three brain regions. These genes may provide a set of potential biomarkers for clinical assessment of drug treatment effectiveness. It is also possible that beneficial effects of HDACi 4b result from alterations in different genes that compensate for deficits in other systems that have been shown to be disrupted in HD. Support for this hypothesis comes from our pathways analysis results, which indicated that HDACi 4b treatment caused a decrease in the expression of genes associated with cell death, cell cycle, and the immune response (Table S1).
Interestingly, the expression of different sets of genes was altered by HDACi 4b treatment in WT mice compared with HD transgenic mice within a given brain region. Furthermore, greater numbers of genes were affected in brains of WT mice compared with HD transgenic mice. This may suggest that mutant htt protein binds to and/or disrupts the nucleosome, resulting in a state of chromatin condensation that cannot be overcome by modifying the acetylation status of histones.
In summary, our findings suggest that therapies aimed at modulating transcription may provide clinical benefits to HD patients, with minimal toxic effects. In particular, we suggest that HDACi 4b treatment may prove useful in slowing the progression of symptoms in humans.
Materials and Methods
Animals.
An R6/2 line of the CBA × C57BL/6 strain origin (Jackson Laboratories) (31) has been maintained at The Scripps Research Institute by breeding male heterozygous R6/2 mice with F1 hybrids of the same background. At the age of 4 weeks, mice were genotyped according to the Jackson Laboratories protocol to determine hemizygosity for the HD transgene. The CAG repeat lengths in these mice were verified by commercial genotyping (Laragen) and found to be close to 300 (291–296); hence, we have designated these mice as R6/2300Q transgenic mice. Characterization of the disease progression in these mice reveals significant rotarod deficits by 12 weeks of age and survival between 6 and 7 months of age.
Drug Treatment.
HDACi 4b [N1-(2-aminophenyl)-N7-phenylheptanediamide] was synthesized as described previously (23). R6/2300Q transgenic mice were individually housed and maintained on a normal 12-h light/dark cycle with lights on at 6:00 a.m. Food and water were available ad libitum. Groups of mice (n = 8 per genotype and drug treatment) were administered HDACi 4b (1 mg/ml) for 67 days in solutions that replaced drinking water beginning at 4 months of age. HDACi 4b was dissolved with HOP-β-CD as an excipient (17); control mice received Arrowhead distilled water containing an equal volume of drug vehicle. The volume of consumed liquid was estimated a few days before the start of treatment, and the concentrations of drug solutions were prepared according to the amount consumed. There were significant differences in the amounts of drug/vehicle solutions consumed per mouse per day across genotype and drug groups (3.66 ± 0.08 ml vs. 3.24 ± 0.11 ml per day for WT vs. transgenic mice, respectively, P = 0.001; 3.62 ± 0.09 ml vs. 3.14 ± 0.12 ml for HDACi 4b- vs. vehicle-treated mice, respectively, P = 0.003). However, given the smaller weights of the transgenic mice, this volume corresponded to approximately the same dose of ≈150 mg/kg/day, most of which was consumed over the 12-h dark cycle. Mice were individually housed to ensure consumption of fluid and food for each mouse. Drug/water consumed was recorded every day, and body weights were recorded two to three times per week. New drug was given every 4–5 days. For microarray experiments, groups of mice (n = 4 per genotype and drug condition) were injected s.c. with HDACi 4b (150 mg/kg/day) or an equal amount of vehicle (50:50, DMSO:PBS) once per day for 3 days. Mice were killed 6 h after the final injection, and brains were removed for microarray processing. All procedures were in strict accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Motor Behavioral Assessments.
Mice were tested on an AccuRotor rotarod (AccuScan Instruments) during the light phase of the 12-h light/dark cycle by using an accelerating rotation paradigm (4–20 rpm). Mice were trained on the rotarod before drug treatment (3 months of age) to establish a behavioral baseline. At baseline, significant differences in rotarod performance are detected in R6/2300Q transgenic mice (288.4 ± 6.3 s vs. 250.0 ± 14.6 s, respectively, P = 0.02). Behavioral semiquantitative assessment of motor and postural abnormalities were scored according to a previously described motor scale (32), which includes the widely used assessment of hindlimb clasping (31), evaluation of truncal dystonia (kyphosis), and general locomotion (including rearing and grooming behavior). Assessments were made by two independent observers who were blind to genotype and treatment status. Details of these tests are provided in SI Methods.
Neuropathologic Evaluations of HDACi 4b Treatment.
At the end of the drug treatment, mice were killed by brief isoflurane exposure and intracardially perfused with cold 4% paraformaldehyde/PBS. After perfusion, brains were weighed and then frozen at −70°C. Striatal and lateral ventricle volumetric analyses were performed on 25-μm brain sections. Sections 125 μm apart spanning the striatum from approximate bregma 1.18–0.38 mm were obtained by microscopic capture using Axiovision LE software (Carl Zeiss). Areas of striatum and lateral ventricle were manually traced from both sides, and volumes were recorded in square millimeters.
Microarray Analysis.
Total RNA was extracted from cortex, striatum, and cerebellum from WT and R6/2300Q transgenic mice treated with vehicle or HDACi 4b. Four replicates were run per sample category, for a total of 48 arrays. RNA quantity was assessed with Nanodrop (Nanodrop Technologies) and quality with the Agilent Bioanalyzer (Agilent Technologies). Total RNA (200 ng) was amplified, biotinylated, and hybridized on Illumina Mouse Mouseref-8 Expression Beadchips v1, querying the expression of ≈22,000 Refseq transcripts, as per the manufacturer's protocol. Slides were scanned by using Illumina BeadStation, and the signal was extracted by using Illumina BeadStudio software. Raw data were analyzed by using Bioconductor packages (33). Quality assessment was performed looking at the interarray Pearson correlation, and clustering based on top variant genes was used to assess overall data coherence. Contrast analysis of differential expression was performed by using the LIMMA package (34). After linear model fitting, a Bayesian estimate of differential expression was calculated. Approximately half of the genes on the array were called Present, resulting in a false discovery rate of 6%. Data analysis was aimed at (i) identifying transcriptional changes in the R6/2300Q transgenic compared with WT mice and (ii) the effect of drug treatment on gene-expression abnormalities. For the initial analyses, the threshold for statistical significance was set at P < 0.005. In a second analysis, we used a less stringent criteria of P < 0.05 for both the disease and drug treatment effects, followed by an absolute log2 ratio of >0.20. Pathways analysis was performed by using the Functional Analysis Annotation tool in Ingenuity Pathways Analysis software (Ingenuity Systems).
Real-Time PCR Experiments.
Real-time PCR experiments were performed as described previously (14) and in SI Methods, using the primer sequences shown in Table S2.
Chromatin Immunoprecipitation.
ChIP assays on brain homogenates were performed as described in previous studies (23). For each immunoprecipitation experiment, the amount of lysate corresponding to 25–50 μg of total DNA was incubated with antibodies directed against histones H3 and H4 or acetylated H3 and H4 (Upstate Biotechnology). The recovered DNA was quantified by real-time PCR analysis using primer pairs directed against promoter regulatory or transcribed regions of the indicated genes (Table S2).
Statistical Analyses.
The data were analyzed with one- or two-factor ANOVA and Student's t tests, using GraphPad Prism Software. For PCR analyses, given that the direction of expression change was predicted a priori from the microarray findings, one-tailed tests were used.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants MH069696 (to E.A.T.) and NS055158 (to J.M.G.), grants from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to G.C. and D.H.G.), and funding from Repligen Corporation.
Footnotes
Conflict of interest statement: J.M.G. acts as a consultant to Repligen Corporation and declares a competing financial interest in this work.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804249105/DCSupplemental.
References
- 1.Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Sugars KL, Rubinsztein DC. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19:233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- 3.Okazawa H. Polyglutamine diseases: A transcription disorder? Cell Mol Life Sci. 2003;60:1427–1439. doi: 10.1007/s00018-003-3013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas EA. Striatal specificity of gene expression dysregulation in Huntington's disease. J Neurosci Res. 2006;84:1151–1164. doi: 10.1002/jnr.21046. [DOI] [PubMed] [Google Scholar]
- 5.Li SH, et al. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol Cell Biol. 2002;22:1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunah AW, et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 7.Luthi-Carter R, et al. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum Mol Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 8.Freiman RN, Tjian R. Neurodegeneration. A glutamine-rich trail leads to transcription factors. Science. 2002;296:2149–2150. doi: 10.1126/science.1073845. [DOI] [PubMed] [Google Scholar]
- 9.Zhai W, et al. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 10.Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 11.Ferrante RJ, et al. Chemotherapy for the brain: The antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington's disease. J Neurosci. 2004;24:10335–10342. doi: 10.1523/JNEUROSCI.2599-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryu H, et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington's disease. Proc Natl Acad Sci USA. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stack EC, et al. Modulation of nucleosome dynamics in Huntington's disease. Hum Mol Genet. 2007;16:1164–1175. doi: 10.1093/hmg/ddm064. [DOI] [PubMed] [Google Scholar]
- 14.Desplats PA, et al. Selective deficits in the expression of striatal-enriched mRNAs in Huntington's disease. J Neurochem. 2006;96:743–757. doi: 10.1111/j.1471-4159.2005.03588.x. [DOI] [PubMed] [Google Scholar]
- 15.Hodges A, et al. Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn A, et al. Mutant huntingtin's effects on striatal gene expression in mice recapitulate changes observed in human Huntington's disease brain and do not differ with mutant huntingtin length or wild-type huntingtin dosage. Hum Mol Genet. 2007;16:1845–1861. doi: 10.1093/hmg/ddm133. [DOI] [PubMed] [Google Scholar]
- 17.Hockly E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci USA. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrante RJ, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington's disease mice. J Neurosci. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardian G, et al. Neuroprotective effects of phenylbutyrate in the N171–82Q transgenic mouse model of Huntington's disease. J Biol Chem. 2005;280:556–563. doi: 10.1074/jbc.M410210200. [DOI] [PubMed] [Google Scholar]
- 20.Kelly WK, O'Connor OA, Marks PA. Histone deacetylase inhibitors: From target to clinical trials. Expert Opin Investig Drugs. 2002;11:1695–1713. doi: 10.1517/13543784.11.12.1695. [DOI] [PubMed] [Google Scholar]
- 21.Dokmanovic M, Marks PA. Prospects: Histone deacetylase inhibitors. J Cell Biochem. 2005;96:293–304. doi: 10.1002/jcb.20532. [DOI] [PubMed] [Google Scholar]
- 22.Drummond DC, et al. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- 23.Herman D, et al. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 24.Rai M, et al. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS ONE. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hockly E, et al. Standardization and statistical approaches to therapeutic trials in the R6/2 mouse. Brain Res Bull. 2003;61:469–479. doi: 10.1016/s0361-9230(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 26.Bates GP, Hockly E. Experimental therapeutics in Huntington's disease: Are models useful for therapeutic trials? Curr Opin Neurol. 2003;16:465–470. doi: 10.1097/01.wco.0000084223.82329.bb. [DOI] [PubMed] [Google Scholar]
- 27.Beckers T, et al. Distinct pharmacological properties of second generation HDAC inhibitors with the benzamide or hydroxamate head group. Int J Cancer. 2007;121:1138–1148. doi: 10.1002/ijc.22751. [DOI] [PubMed] [Google Scholar]
- 28.Fennema-Notestine C, et al. In vivo evidence of cerebellar atrophy and cerebral white matter loss in Huntington disease. Neurology. 2004;63:989–995. doi: 10.1212/01.wnl.0000138434.68093.67. [DOI] [PubMed] [Google Scholar]
- 29.Broide RS, et al. Distribution of histone deacetylases 1–11 in the rat brain. J Mol Neurosci. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- 30.Sadri-Vakili G, et al. Histones associated with downregulated genes are hypo-acetylated in Huntington's disease models. Hum Mol Genet. 2007;16:1293–1306. doi: 10.1093/hmg/ddm078. [DOI] [PubMed] [Google Scholar]
- 31.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 32.Fernagut PO, et al. Subacute systemic 3-nitropropionic acid intoxication induces a distinct motor disorder in adult C57Bl/6 mice: Behavioural and histopathological characterisation. Neuroscience. 2002;114:1005–1017. doi: 10.1016/s0306-4522(02)00205-1. [DOI] [PubMed] [Google Scholar]
- 33.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, et al., editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.