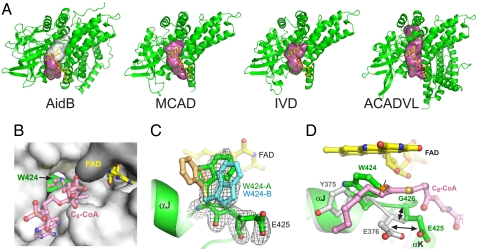
Fig. 3.
The FAD and putative substrate binding pocket. (A) Comparison of the substrate access cavities (magenta/white surfaces) in AidB, MCAD, IVD, and ACADVL (very long chain acyl-CoA dehydrogenase, PDB ID code 2UXW). The rear of the cavity blocked by Trp-424 in AidB is shown as a white surface. (B) The substrate access channel as viewed from the outside of the protein (gray surface). Yellow sticks, FAD; green, Trp-424. The octanoyl-CoA substrate (pink sticks) from the MCAD structure (PDB ID code 3MDE) is superimposed for reference. (C) Annealed omit electron density (3σ contours) for Trp-424 and Glu-425. The two Trp-424 conformers modeled into the density are shown as green and blue sticks. A hypothetical conformer (tan sticks) in the same position as Tyr-375 in ACAD structures is shown for reference only and was not included in the final structure. (D) Superposition of AidB (green) and MCAD (gray) structures. The AidB flavin is yellow, and the MCAD octanoyl-CoA substrate is pink. The black arrows highlight the shift in Glu-425 position, and the steric clash between Trp-424 and a fatty acyl substrate is shown as an orange starburst.