Abstract
The cross-linked (cornified) envelope is a characteristic product of terminal differentiation in the keratinocyte of the epidermis and related epithelia. This envelope contains many proteins of which involucrin was the first to be discovered and shown to become cross-linked by a cellular transglutaminase. Involucrin has evolved greatly in placental mammals, but retains the glutamine repeats that make it a good substrate for the transglutaminase. Until recently, it has been impossible to detect involucrin outside the placental mammals, but analysis of the GenBank and Ensembl databases that have become available since 2006 reveals the existence of involucrin in marsupials and birds. We describe here the properties of these involucrins and the ancient history of their evolution.
Keywords: aves, marsupial, evolution, glutamine repeats
The outer surface of the skin of humans and other mammals consists of dead cells, each of which contains a chemically resistant envelope. The nature of this envelope has been extensively reviewed (1, 2). Envelopes formed in cultures of epidermal cells are insoluble in ionic detergents at 100°C, but are dissolved by proteolytic enzymes (3). The envelopes are ≈120 Å in thickness (4) and composed of proteins heavily cross-linked by ε-(γ glutamyl) lysine (isopeptide) bonds introduced by the action of transglutaminase (5). A protein ultimately incorporated into the cross-linked envelope was discovered as a soluble precursor before the activation of the cross-linking (6, 7). This precursor was named involucrin (from the Latin for envelope: involucrum) and was present only in enlarging cells undergoing terminal differentiation (8, 9). Because involucrin is a substrate of transglutaminase, it is not surprising that it contains numerous glutamines capable of participating in the cross-linking reaction. Human involucrin contains 38–42 repeats of a 10 amino acid sequence, each repeat containing 3 glutamine residues (10–13).
Other protein precursors of the cross-linked envelope were soon discovered (14). One had a molecular mass of 210 kDa and was later named envoplakin (15). Another had a molecular mass of 195 kDa and was later named periplakin (16). Still other precursors were discovered, including filaggrin (17), small proline-rich repeat proteins or SPRRs (18), and loricrin (19). The genes encoding most envelope precursors (but not periplakin or envoplakin) are located in the region of human chromosome 1q21, the so-called EDC or epidermal differentiation complex (20). This has been shown for involucrin (13), filaggrin (21), loricrin (22), cornifin (23), the SPRRs (24), and others (25). Other proteins encoded in the EDC, such as cornulin (26), appear late in terminal differentiation, but are not envelope precursors. Still others are precursors that are incorporated after the envelope is formed, particularly the “late envelope proteins” (27). It has been proposed that because involucrin, loricrin, and SPRR proteins have similar amino acid sequences in their N-terminal and C-terminal domains, they probably originated by successive duplications of a single ancestral gene, followed by the divergence of each gene (28, 29).
Owing to the rapidity of evolutionary change in involucrin (29), there are important differences between the involucrins of primates and nonprimate mammals. Antibodies to mammalian involucrin do not detect involucrin in taxa below the placental mammals. Similarly, because of sequence divergence in the gene, cDNA that encodes mammalian involucrin does not detect involucrin mRNA in lower taxa. From the study of GenBank data, it has become possible to identify involucrin in classes outside the mammals. The identification of these extra-mammalian involucrins depends on the fact that the gene order in the EDCs of remote species has been largely retained.
Results
Detection of Envelope Precursor Genes in the Marsupial Monodelphis domestica (the Opossum).
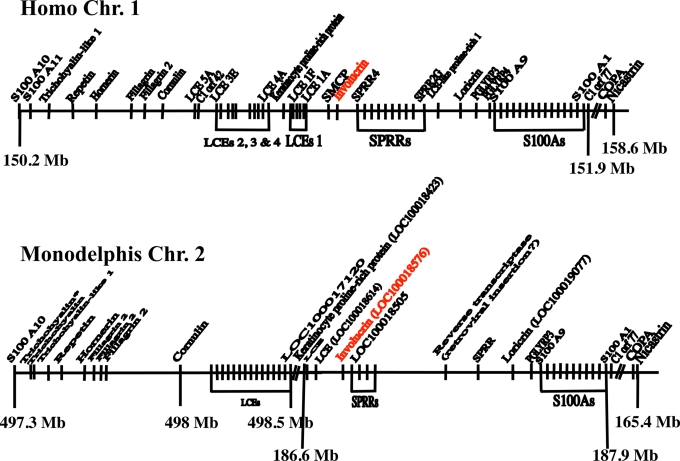
To identify the involucrin gene of Monodelphis, a similarity search of the Monodelphis genomic sequences deposited at the Ensembl database (www.ensembl.org) was carried out by using the entire mouse involucrin sequence. The ab initio§ peptides of the Ensembl database were searched with the BLASTP program by using default parameters (nearly exact matches). The greatest similarity was obtained between residues 175–314 of mouse involucrin and a putative peptide encoded by the region of Monodelphis chromosome 2 located at coordinate 186.8 Mb from the tip of the short arm. Examination of this region in GenBank disclosed a continuous ORF (LOC100018576) beginning with an ATG codon and encoding a 326-residue protein (including the initiating methionine). This locus is surrounded by LOC100018614 and LOC100018505, which were found to encode putative proteins similar to a late cornified envelope protein (LCE) and to an SPRR, respectively (Fig. 1). It is worth noting that the GenBank (American) and Ensembl (European) databases both contain a Monodelphis entry labeled “similar to involucrin,” but neither of these two putative proteins is likely to be involucrin. GenBank transcript XM_001364369 derived from gene LOC100010894, whose chromosomal location is unknown, encodes a proline-rich protein whose amino acid composition is very different from that of involucrin. Ensembl transcript ENSMODG00000018957 is derived from gene LOC100019299, which is located in chromosome 2 at coordinate 497.36 Mb, within the trichohyalin gene cluster. In keeping with its sequence and the position of the gene, its product is likely to be a member of the trichohyalin family. Because in both human and mouse the nearest neighbors of the involucrin gene are genes encoding an LCE and an SPRR, it seemed likely that LOC100018576 of Monodelphis chromosome 2 is the authentic involucrin gene.
Fig. 1.
The human and Monodelphis EDC regions. The human EDC extends over 1.7 Mb and is bounded at its 5′ end by S100A10 and at its 3′ end by C1 orf77, COPA, and nicastrin. Because these two termini are extremely conserved in evolution, they could be used to define the boundaries of the EDC in Monodelphis. Both termini were found on Monodelphis Chromosome 2, but they were separated by approximately 300 Mb instead of the 1.7 Mb of the human. This was because of the insertion of large regions syntenic to human chromosomes 17 (≈100 Mb) and 6 (≈200 Mb) near the middle of the Monodelphis gene. The point of interruption of the Monodelphis EDC could be determined from the gene map of chromosome 2 in GenBank. It is located between one of the LCE genes (LOC100017120) and the keratinocyte proline-rich protein gene (LOC100018423). Because some of the genes in the GenBank map were not identified, we carried out BLASTN identity searches to identify them. We found that in Monodelphis, a rearrangement occurred such that the order of the two halves was reversed. The effect of this rearrangement has been eliminated on the figure by again reversing the two halves to align the sequence of Monodelphis with that of the human.
Whereas the sequence of the Monodelphis involucrin gene diverges considerably from that of the human involucrin gene, the evidence that it indeed encodes involucrin may be summarized as follows:
The coding region is confined to a single exon.
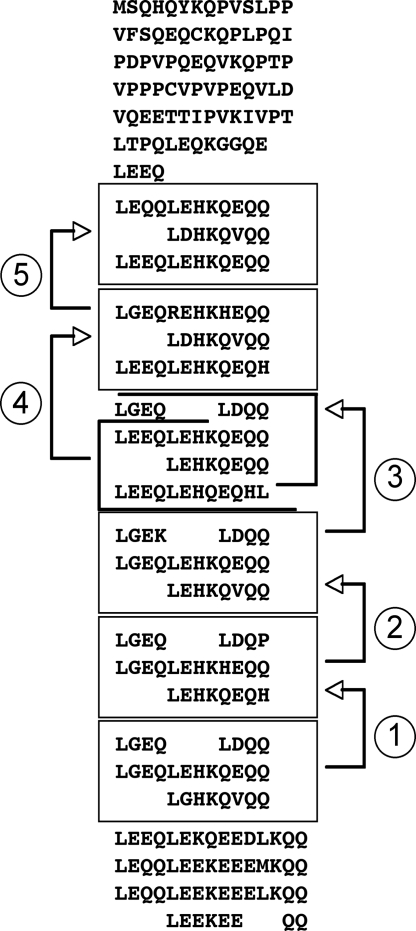
The Monodelphis gene encodes repeats that, although somewhat irregular (8–12 amino acids), appear to have resulted from successive duplications of blocks of three repeats (Fig. 2). In the average repeat, approximately 25% of the codons encode Q and another 30% differ from a glutamine codon by a single nucleotide substitution.
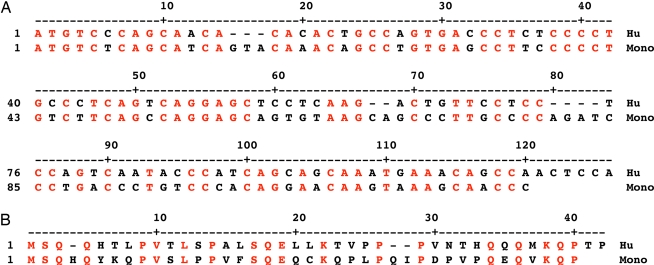
The distance between the initiating methionine and the start of the repeats in Monodelphis is nearly identical to that of the nonprimate mammals (82 codons). A sequence matching program reveals extensive identity of nucleotide sequence and encoded amino acids of the two species (Fig. 3).
As in the placental mammals, the Monodelphis involucrin gene is located immediately upstream of the SPRR genes (Fig. 1).
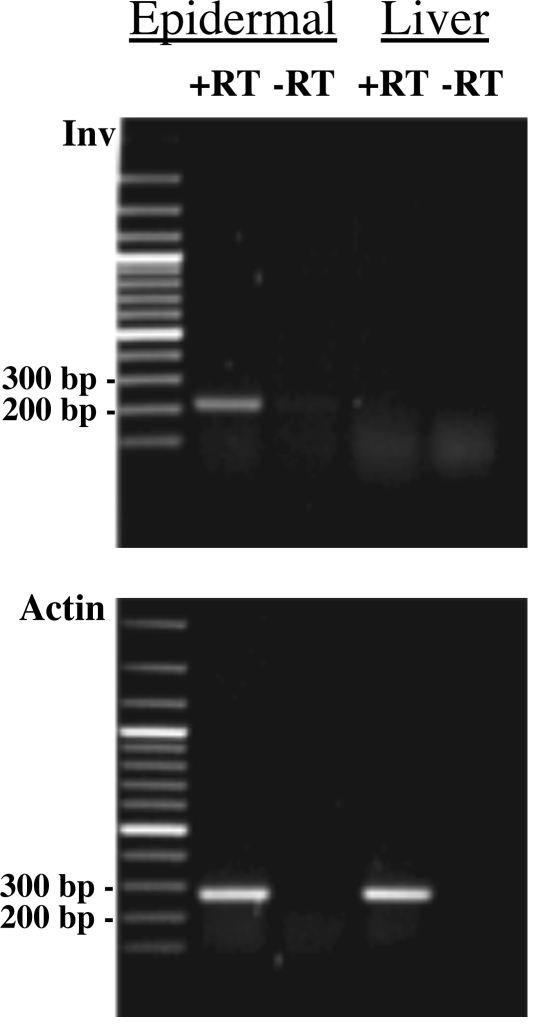
The mRNA encoding Monodelphis involucrin was easily detected in the epidermis of the animal by RT-PCR and was absent from liver, an organ that does not contain stratified squamous epithelium (Fig. 4).
Fig. 2.
Involucrin of Monodelphis. A series of five duplications (arrows 1–5) of a block of three repeats would create most of the involucrin of Monodelphis domestica.
Fig. 3.
Alignment of Human and Monodelphis 5′ end of involucrin coding exon. (A) Nucleotide sequence. (B) Encoded amino acid sequence. This was carried out by using the MegAlign program of DNAstar Lasergene v6 (courtesy of S. Iuchi, Harvard Medical School). There is similarity of nucleotide sequence and conservation of numerous encoded amino acids (in red type).
Fig. 4.
RT-PCR on RNA prepared from epidermis and liver of Monodelphis domestica. Epidermis of Monodelphis was separated from dermis with thermolysin. RNA was prepared and RT-PCR was carried out by using involucrin sense and antisense primers whose sequences were 5′ ATG TCT CAG CAT CAG TAC AAA and 5′ AGT AGG AAC TAT CTT CAC AGG, respectively. The product was the result of 30 cycles of amplification (1 min, 95°; 1 min, 57°; and 1 min, 72°). Expected size of band: 207 bp. A band of expected size is visible in the presence of epidermal RNA and reverse transcriptase (RT). No band is visible in the absence of RT. No involucrin transcript is detected in liver. Similar amounts of actin mRNA were detected in epidermis and liver. The gene for the putative involucrin of Monodelphis is therefore expressed in the epidermis, but not in the liver.
Because Monodelphis, like the placental mammals, possesses an EDC region including involucrin, the history of the involucrin gene is older than the eutherian–metatherian divergence, which is believed to have occurred in the Late Cretaceous Period, approximately 125–147 million years ago. Like the involucrin gene of placental mammals, the marsupial involucrin gene has evolved by successive repeat addition. The marsupial gene contains the segment of repeats at site P, identified many years ago in the nonanthropoid placental mammals (30, 31). Therefore, this segment of repeats must have been generated in an ancestor of the placental mammals and the marsupials.
Detection of Envelope Precursor Genes in Aves (Gallus gallus).
The availability of GenBank and Ensembl data has also permitted the detection of involucrin in Aves, a group much more remote from the mammals than are the marsupials. Sauropsids, the monophyletic group that includes birds and reptiles, diverged from the mammalian lineage in the last half of the Carboniferous Period, >300 million years ago (32).
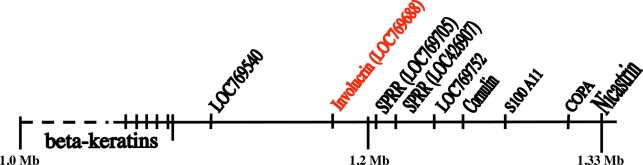
The genes for COPA, nicastrin, and S100A11 are clustered toward the 3′ end of the human and Monodelphis EDCs. All three genes are very conserved in evolution and could therefore be used to localize the Gallus EDC. BLASTN identity searches of the chicken genomic sequences in the Ensembl database disclosed that the genes for COPA, nicastrin, and S100A11 were clustered on chromosome 25 at approximately 1.3 Mb (Fig. 5). We assumed that this position represented the 3′ end of the Gallus EDC. This was confirmed by the fact that upstream of the S100A11 gene we could locate two SPRR genes (LOC426907 and LOC769705). As in the human and Monodelphis, the involucrin gene should lie immediately upstream of the upstream-most SPRR gene. At this position (LOC769688), we indeed found the Gallus involucrin gene.
Fig. 5.
The EDC region of Gallus on chromosome 25. Unexpectedly, this region is linked to the genes for beta keratins, which are specific to sauropsids and might be derived from the ancestral gene of the EDC.
The amino acid sequence of the involucrin gene of Gallus is compared with that of the human in Fig. 6. The 463 amino acids encoded in the Gallus gene have numerous matches with the 585 amino acids encoded in the human gene. Most matches are of glutamines, including numerous doublets and a single triplet, and there are also matches of prolines and occasionally of lysines, but there are no reiterations of these amino acids.
Fig. 6.
Alignment of amino acid sequence of human and Gallus involucrins. Identities shown in red type.
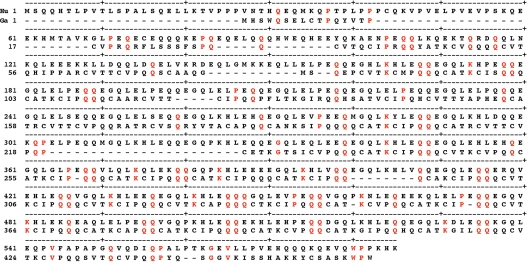
The 3′-most repeat of the Gallus gene is located at amino acids 428–436 of the sequence of Fig. 6. The Gallus sequence contains a total of 43 glutamine-rich repeats, each usually beginning with PQQ and ending with a hydrophobic residue (Fig. 7). The most specific property of all involucrins is that each of its repeats contains 2–4 consecutive glutamines. The repeats of the putative 463-residue protein encoded by LOC769688 most commonly contain 10 amino acids, of which three are consecutive glutamines (Fig. 7). These repeats do not have the regularity of the repeats of mammalian involucrins, but they have many features in common. In both taxa the repeats have undergone successive duplications. One block of five repeats in Gallus has been almost exactly duplicated twice (Fig. 7).
Fig. 7.
Repeat structure of Gallus involucrin (GenBank XP_001232979). Of a total of 43 repeats, 36 have 9–12 amino acids, 21 contain 10 amino acids, usually including three glutamines. A block of five repeats has been almost exactly duplicated twice (arrows 1 and 2).
Although other proteins encoded in the EDC are commonly composed of repeats. These repeats, unlike those of involucrin, do not contain reiterated glutamines.
Detection of Involucrin in Cultured Cells and Epidermis of Gallus.
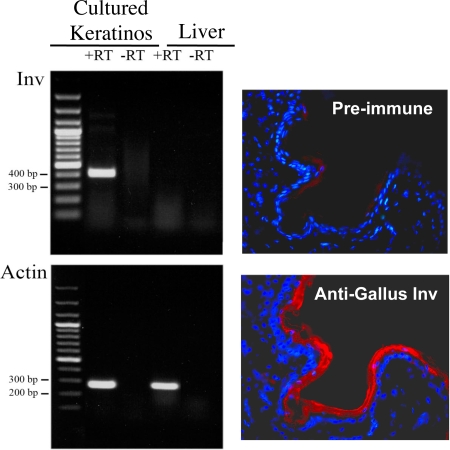
Because the putative involucrin gene that we had found had been predicted by computer algorithms only, we decided to obtain experimental evidence supporting transcription of the gene and existence of the encoded protein. Newborn-chicken keratinocytes were cultivated as described in ref. 33. When the cells were confluent, we prepared total RNA and carried out an RT-PCR specified for the predicted chicken involucrin mRNA. This analysis revealed the existence of an abundant product of the expected size in keratinocytes of the chicken, but not in its liver (Fig. 8).
Fig. 8.
Experimental verification of GenBank data. (Left) RT-PCR of Gallus mRNA by using cultured chicken epidermal cells. Cultures were prepared (33) and RNA was isolated. RT-PCR was carried out by using primers 5′ CTGGCAGTCAGAGCTGTGCAC (sense) and 5′ GGCGGATTCCCTTGGTCAGGAAT (antisense) with 30 cycles. A band of the expected size (392 bp) was obtained. A control of liver gave no band. An actin band was obtained from both cultured keratinocytes and liver. (Right) Detection of Gallus involucrin with an anti-peptide antibody. Antibodies to Gallus involucrin were prepared by Primm. A section through Gallus skin was stained with the antiserum (red) and counterstained with DAPI (blue) for DNA. The cornified layer of the epidermis was strongly stained. The preimmune serum gave only faint patchy background staining.
To demonstrate the existence of the protein encoded by LOC769688, a polyclonal antiserum was prepared. A mixture of three synthetic peptides was used for immunization. The peptide sequences were PRQQYATKCVQQ, VTTYAPHEQCATR, and KISSHAKKYCSASK corresponding to codons 39–50, 147–159, and 447–460. The antiserum detected the protein encoded by LOC769688 in sections of epidermis of newborn chicken. The protein had the typical distribution in the outer layers (Fig. 8). No staining was detected in the fibroblasts of the dermis. We may conclude that the protein encoded by LOC769688 is indeed involucrin.
Discussion
The use of GenBank and Ensembl data has made possible the identification of the involucrin genes of marsupials and birds, a discovery that could not have been made by previously available methods. The resemblances between the repeat structures of the involucrin genes of those taxa and those of mammals are evident. Moreover, the existence of the Gallus gene has been confirmed experimentally by showing that it is expressed in cultured Gallus epidermal cells, but not in other cell types, and that an antibody to a peptide sequence revealed by the GenBank data detected involucrin in epidermis. In this way it has been possible to verify the correctness of the GenBank and Ensembl data. It has also been possible to correct errors of interpretation based on gene-prediction algorithms alone.
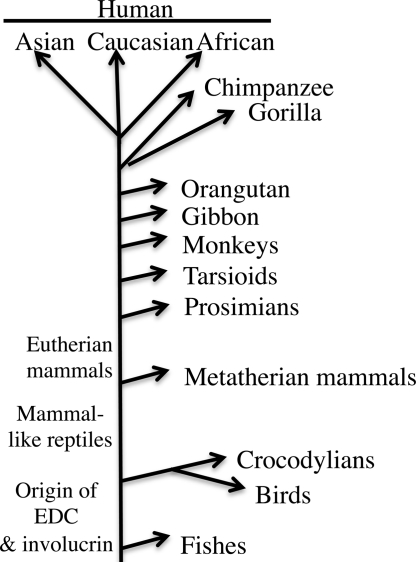
The existence of an involucrin gene of Gallus indicates that the gene was present in a common ancestor of birds and mammals >300 million years ago. The process of repeat addition has been occuring since that time and is no doubt continuing in mammals today, as shown by the existence of many involucrin polymorphisms in human and other mammalian populations (11, 12, 34–39). It is not yet known how long ago in evolution involucrin and the EDC originated. Both may be present in any species possessing a stratified squamous epithelium. We were unable to find an EDC in Danio (a genus that includes the zebrafish), whose sequence is complete. The case of Xenopus is less clear because the sequencing is not very advanced. The evolution of involucrin is summarized in Fig. 9.
Fig. 9.
Evolution of involucrin. For detailed analysis of the later stages of this tree, see refs 11, 12, 29, and 41.
The evolutionary persistence of involucrin and its continuing repeat additions are astonishing, in view of the fact that ablation of this gene in the mouse produces no detectable phenotype. Mice lacking involucrin reproduce normally and have a normal lifespan. Their epidermis appears normal in its structure, heals normally after wounding, and contains cornified envelopes indistinguishable from those of the wild-type mice. No difference can be detected in resistance to physical or chemical agents between the envelopes lacking involucrin and wild-type envelopes (40). This offers no support for an explanation of the evolution of involucrin based on natural selection.
Acknowledgments.
This work was supported by the Centre National de la Recherche Scientifique, the Association pour la Recherche sur le Cancer, and the Ligue contre le Cancer. A.V. was a recipient of fellowships from the Fondation pour la Recherche Médicale and the Fondation Bettencourt–Schueller.
Footnotes
The authors declare no conflict of interest.
All ab initio predictions are solely based on the genomic sequence and do not exploit any other experimental evidence. Therefore, not all translations of predicted transcripts represent real proteins. Consequently these predictions should be used with care.
References
- 1.Hohl D. Cornified cell envelope. Dermatologica. 1990;180:201–211. doi: 10.1159/000248031. [DOI] [PubMed] [Google Scholar]
- 2.Reichert U, Michel S, Schmidt R. In: Molecular Biology of the Skin: The Keratinocyte. Darmon M, Blumenberg M, editors. San Diego, CA: Academic; 1993. pp. 107–140. [Google Scholar]
- 3.Sun TT, Green H. Differentiation of the epidermal keratinocyte in cell culture: Formation of the cornified envelope. Cell. 1976;9:511–521. doi: 10.1016/0092-8674(76)90033-7. [DOI] [PubMed] [Google Scholar]
- 4.Green H. Terminal differentiation of cultured human epidermal cells. Cell. 1977;11:405–416. doi: 10.1016/0092-8674(77)90058-7. [DOI] [PubMed] [Google Scholar]
- 5.Rice RH, Green H. The cornified envelope of terminally differentiated human epidermal keratinocytes consists of cross-linked protein. Cell. 1977;11:417–422. doi: 10.1016/0092-8674(77)90059-9. [DOI] [PubMed] [Google Scholar]
- 6.Rice RH, Green H. Relation of protein synthesis and transglutaminase activity to formation of the cross-linked envelope during terminal differentiation of the cultured human epidermal keratinocyte. J Cell Biol. 1978;76:705–711. doi: 10.1083/jcb.76.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice RH, Green H. Presence in human epidermal cells of a soluble protein precursor of the cross-linked envelope: Activation of the cross-linking by calcium ions. Cell. 1979;18:681–694. doi: 10.1016/0092-8674(79)90123-5. [DOI] [PubMed] [Google Scholar]
- 8.Watt FM, Green H. Involucrin synthesis is correlated with cell size in human epidermal cultures. J Cell Biol. 1981;90:738–742. doi: 10.1083/jcb.90.3.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watt FM, Green H. Stratification and terminal differentiation of cultured epidermal cells. Nature. 1982;295:434–436. doi: 10.1038/295434a0. [DOI] [PubMed] [Google Scholar]
- 10.Eckert RL, Green H. Structure and evolution of the human involucrin gene. Cell. 1986;46:583–589. doi: 10.1016/0092-8674(86)90884-6. [DOI] [PubMed] [Google Scholar]
- 11.Djian P, Delhomme B, Green H. Origin of the polymorphism of the involucrin gene in Asians. Am J Hum Genet. 1995;56:1367–1372. [PMC free article] [PubMed] [Google Scholar]
- 12.Simon M, Phillips M, Green H. Polymorphism due to variable number of repeats in the human involucrin gene. Genomics. 1991;9:576–580. doi: 10.1016/0888-7543(91)90349-j. [DOI] [PubMed] [Google Scholar]
- 13.Simon M, et al. Absence of a single repeat from the coding region of the human involucrin gene leading to RFLP. Am J Hum Genet. 1989;45:910–916. [PMC free article] [PubMed] [Google Scholar]
- 14.Simon M, Green H. Participation of membrane-associated proteins in the formation of the cross-linked envelope of the keratinocyte. Cell. 1984;36:827–834. doi: 10.1016/0092-8674(84)90032-1. [DOI] [PubMed] [Google Scholar]
- 15.Ruhrberg C, et al. Envoplakin, a novel precursor of the cornified envelope that has homology to desmoplakin. J Cell Biol. 1996;134:715–729. doi: 10.1083/jcb.134.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruhrberg C, Hajibagheri MA, Parry DA, Watt FM. Periplakin, a novel component of cornified envelopes and desmosomes that belongs to the plakin family and forms complexes with envoplakin. J Cell Biol. 1997;139:1835–1849. doi: 10.1083/jcb.139.7.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, et al. Evidence for filaggrin as a component of the cell envelope of the newborn rat. Biochem J. 1988;253:153–160. doi: 10.1042/bj2530153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kartasova T, van de Putte P. Isolation, characterization, and UV-stimulated expression of two families of genes encoding polypeptides of related structure in human epidermal keratinocytes. Mol Cell Biol. 1988;8:2195–2203. doi: 10.1128/mcb.8.5.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehrel T, et al. Identification of a major keratinocyte cell envelope protein, loricrin. Cell. 1990;61:1103–1112. doi: 10.1016/0092-8674(90)90073-n. [DOI] [PubMed] [Google Scholar]
- 20.Mischke D, et al. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (“epidermal differentiation complex”) on human chromosome 1q21. J Invest Dermatol. 1996;106:989–992. doi: 10.1111/1523-1747.ep12338501. [DOI] [PubMed] [Google Scholar]
- 21.McKinley-Grant LJ, et al. Characterization of a cDNA clone encoding human filaggrin and localization of the gene to chromosome region 1q21. Proc Natl Acad Sci USA. 1989;86:4848–4852. doi: 10.1073/pnas.86.13.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoneda K, et al. The human loricrin gene. J Biol Chem. 1992;267:18060–18066. [PubMed] [Google Scholar]
- 23.Marvin KW, et al. Cornifin, a cross-linked envelope precursor in keratinocytes that is down-regulated by retinoids. Proc Natl Acad Sci USA. 1992;89:11026–11030. doi: 10.1073/pnas.89.22.11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hohl D, et al. The small proline-rich proteins constitute a multigene family of differentially regulated cornified cell envelope precursor proteins. J Invest Dermatol. 1995;104:902–909. doi: 10.1111/1523-1747.ep12606176. [DOI] [PubMed] [Google Scholar]
- 25.Volz A, et al. Physical mapping of a functional cluster of epidermal differentiation genes on chromosome 1q21. Genomics. 1993;18:92–99. doi: 10.1006/geno.1993.1430. [DOI] [PubMed] [Google Scholar]
- 26.Contzler R, Favre B, Huber M, Hohl D. Cornulin, a new member of the “fused gene” family, is expressed during epidermal differentiation. J Invest Dermatol. 2005;124:990–997. doi: 10.1111/j.0022-202X.2005.23694.x. [DOI] [PubMed] [Google Scholar]
- 27.Marshall D, Hardman MJ, Nield KM, Byrne C. Differentially expressed late constituents of the epidermal cornified envelope. Proc Natl Acad Sci USA. 2001;98:13031–13036. doi: 10.1073/pnas.231489198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Backendorf C, Hohl D. A common origin for cornified envelope proteins? Nat Genet. 1992;2:91. doi: 10.1038/ng1092-91. [DOI] [PubMed] [Google Scholar]
- 29.Green H, Djian P. Consecutive actions of different gene-altering mechanisms in the evolution of involucrin. Mol Biol Evol. 1992;9:977–1017. doi: 10.1093/oxfordjournals.molbev.a040775. [DOI] [PubMed] [Google Scholar]
- 30.Phillips M, Djian P, Green H. The involucrin gene of the galago. Existence of a correction process acting on its segment of repeats. J Biol Chem. 1990;265:7804–7807. [PubMed] [Google Scholar]
- 31.Tseng H, Green H. Remodeling of the involucrin gene during primate evolution. Cell. 1988;54:491–496. doi: 10.1016/0092-8674(88)90070-0. [DOI] [PubMed] [Google Scholar]
- 32.Spinar ZV. Life Before Man. New York: Thames and Hudson, Inc.; 1995. [Google Scholar]
- 33.Vanhoutteghem A, Londero T, Ghinea N, Djian P. Serial cultivation of chicken keratinocytes, a composite cell type that accumulates lipids and synthesizes a novel beta-keratin. Differentiation. 2004;72:123–137. doi: 10.1111/j.1432-0436.2004.07204002.x. [DOI] [PubMed] [Google Scholar]
- 34.Delhomme B, Djian P. Expansion of mouse involucrin by intra-allelic repeat addition. Gene. 2000;252:195–207. doi: 10.1016/s0378-1119(00)00237-7. [DOI] [PubMed] [Google Scholar]
- 35.Djian P, Delhomme B. Systematic repeat addition at a precise location in the coding region of the involucrin gene of wild mice reveals their phylogeny. Genetics. 2005;169:2199–2208. doi: 10.1534/genetics.104.036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Djian P, Green H. Involucrin gene of tarsioids and other primates: Alternatives in evolution of the segment of repeats. Proc Natl Acad Sci USA. 1991;88:5321–5325. doi: 10.1073/pnas.88.12.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teumer J, Green H. Divergent evolution of part of the involucrin gene in the hominoids: Unique intragenic duplications in the gorilla and human. Proc Natl Acad Sci USA. 1989;86:1283–1286. doi: 10.1073/pnas.86.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tseng H, Green H. The involucrin gene of the owl monkey: Origin of the early region. Mol Biol Evol. 1989;6:460–468. doi: 10.1093/oxfordjournals.molbev.a040563. [DOI] [PubMed] [Google Scholar]
- 39.Parenteau NL, Eckert RL, Rice RH. Primate involucrins: Antigenic relatedness and detection of multiple forms. Proc Natl Acad Sci USA. 1987;84:7571–7575. doi: 10.1073/pnas.84.21.7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Djian P, Easley K, Green H. Targeted ablation of the murine involucrin gene. J Cell Biol. 2000;151:381–388. doi: 10.1083/jcb.151.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Urquhart A, Gill P. Tandem-repeat internal mapping (TRIM) of the involucrin gene: Repeat number and repeat-pattern polymorphism within a coding region in human populations. Am J Hum Genet. 1993;53:279–286. [PMC free article] [PubMed] [Google Scholar]