Abstract
Marfan syndrome (MFS), a relatively common autosomal dominant hereditary disorder of connective tissue with prominent manifestations in the skeletal, ocular, and cardiovascular systems, is caused by mutations in the gene for fibrillin‐1 (FBN1). The leading cause of premature death in untreated individuals with MFS is acute aortic dissection, which often follows a period of progressive dilatation of the ascending aorta. Recent research on the molecular physiology of fibrillin and the pathophysiology of MFS and related disorders has changed our understanding of this disorder by demonstrating changes in growth factor signalling and in matrix‐cell interactions. The purpose of this review is to provide a comprehensive overview of recent advances in the molecular biology of fibrillin and fibrillin‐rich microfibrils. Mutations in FBN1 and other genes found in MFS and related disorders will be discussed, and novel concepts concerning the complex and multiple mechanisms of the pathogenesis of MFS will be explained.
Keywords: fibrillin, Marfan syndrome, microfibril, TGFβ
Marfan syndrome (MFS; MIM 154700) is a relatively common autosomal dominant hereditary disorder of connective tissue with prominent manifestations in the skeletal, ocular, and cardiovascular systems. MFS is caused by mutations in the gene for fibrillin‐1 (FBN1). Many affected individuals have a characteristic habitus with tall stature, long slender limbs (dolichostenomelia), arachnodactyly, scoliosis, and pectus excavatum or carinatum. Ectopia lentis affects up to 80% of individuals with MFS and is almost always bilateral. The leading cause of premature death in untreated individuals with MFS is acute aortic dissection, which follows a period of progressive dilatation of the ascending aorta. Recent comprehensive treatments of the clinical aspects of MFS have been published.1,2
A review of the molecular genetics of MFS appeared in these pages in the year 2000.3 In the half decade since then, remarkable progress has been made in elucidating the molecular anatomy of both fibrillin and the fibrillin‐rich microfibrils as well as in understanding the molecular pathogenesis of MFS. Initial ideas about the pathogenesis of MFS concentrated on a static dominant negative model based on the concept of fibrillin‐rich microfibrils as purely architectural elements in the extracellular matrix. Recent findings greatly enhanced our understanding of the pathogenesis of MFS by demonstrating changes in growth factor signalling and other changes in matrix‐cell interactions, which has set the stage for attempts to develop novel forms of treatment.
The current review will focus on the fibrillin‐LTBP gene family and tissue organisation of microfibrils, mutations in genes associated with MFS and related phenotypes, the structure of fibrillin‐1 domains, FBN1 mutations and proteolysis, and the genetics of MFS in mouse models. The current review will highlight advances published after the first review; interested readers are referred to the previous article for more information on other topics.3
The fibrillin‐LTBP gene family
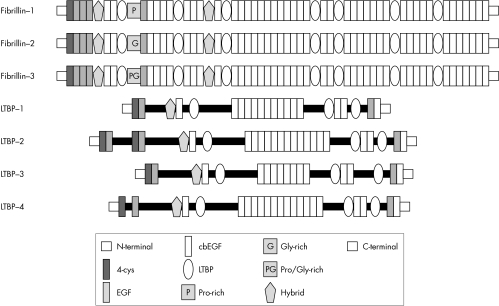
The fibrillins and the latent‐TGFβ‐binding proteins (LTBPs) form two closely related protein families with structural and non‐structural functions in the extracellular matrix. Both families are characterised by a modular domain structure with repeated cysteine‐rich modules.
Members of the fibrillin‐LTBP family have both structural and non‐structural functions. Three closely related fibrillins have been described. Fibrillin‐24 and the recently discovered fibrillin‐35 have a domain organisation identical to that of fibrillin‐1 and an overall level of amino acid identity of between 61% and 69%. Several lines of evidence suggest that the fibrillins have both overlapping and unique functions.5,6 All three fibrillins are structural components of microfibrils4,5,7; however, fibrillin‐2 and fibrillin‐3 are preferentially expressed in embryonic developmental stages,4,5,8 whereas fibrillin‐1 is expressed from the gastrula to throughout adult life.9 In addition, distinct phenotypes are observed in fbn1 and fbn2 gene targeting experiments in mice; since microfibrils are assembled in both mice, some overlap of architectural functions appears likely.5,10,11,12
The LTBPs associate with transforming growth factor‐β (TGFβ), thereby regulating its secretion and spatial and temporal activation; in humans, four members of the LTBP family are known, three of which undergo alternative splicing.13 TGFβ is synthesised as a homodimeric proprotein, whereby the dimeric propeptide is cleaved intracellularly from the growth factor; the propeptide is called the latency‐associated peptide (LAP) because TGFβ cannot bind to its surface receptors when it is bound to the LAP. The LAP in turn is usually disulfide‐bonded to an LTBP; this aggregate is referred to as the large latent complex (LLC). The LTBPs thus have a dual function: as structural components of the extracellular matrix and as modulators of TGFβ availability (the reader is referred to Rifkin14 and Todorovic et al15 for recent reviews on this topic).
The fibrillins and LTBPs display a remarkably similar domain structure made up of repeated cysteine‐rich structural modules including multiple copies of an epidermal growth factor‐like module (EGF) and an 8‐cysteine (8‐Cys) module found only in the fibrillins and LTBPs (fig 1). EGF modules are approximately 45 residues in length and are characterised by six conserved cysteine residues that form three intramodule disulfide bonds. Forty three of the 47 EGF modules in fibrillin‐1 and many of those in the various LTBPs additionally conform to a consensus sequence, (D/N)‐X‐(D/N)(E/Q)Xm(D/N)*Xn(Y/F), that mediates calcium binding in the N‐terminal region of the module (m and n are variable, and an asterisk denotes β‐hydroxylation).16
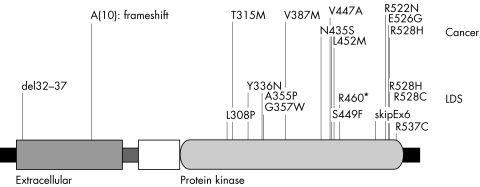
Figure 2 Mutations in TGFBR2 found in Loeys‐Dietz aortic aneurysm syndrome (LDS)112 and related hereditary disorders111,113 and representative missense mutations found in colon carcinoma166,250 and breast carcinoma.251 The TGFBR2 protein is drawn according to UniProt entry P37173 and comprises an N‐terminal signal sequence, an extracellular domain, a transmembrane domain (stippled rectangle), a cytoplasmatic region of unknown significance (the white box), the serine‐threonine protein kinase domain, and a C‐terminal domain (black rectangle).
Figure 1 The fibrillin‐1 gene FBN1 spans about 235 kb of genomic DNA on chromosome 15q21.1, and has a transcript size of 9749 nucleotides; the coding sequence of FBN1 is spread over 65 exons,21 and three alternatively spliced non‐coding 5′ exons have been described.249 With several exceptions, single exons code for the domains in fibrillin‐1 as shown here.
The 8‐Cys module (also variously referred to in the literature as the 8‐cysteine module, the TB module, or the LTBP module) occurs only in the fibrillins and LTBPs. An experiment using a recombinant construct of one of the seven 8‐Cys modules of fibrillin‐1 showed that the structure is stabilised by four intradomain disulfide bonds.17 Some 8‐Cys domains can mediate binding to the LAP‐TGFβ complex.18,19 Not all domains have this function; a recent study showed that LTBP1 and LTBP3 bound efficiently to TGFβ, LTBP4 bound weakly, and LTBP2 and fibrillin‐1 and fibrillin‐2 did not bind.20 An additional cysteine‐rich module with similarities to both the cbEGF module and the 8‐Cys module, termed the 8‐Cys hybrid module,21 can mediate intermolecular disulfide bonding between fibrillin‐1 monomers, which may be an important step in the assembly of microfibrils.22 The fibrillins share a globular C‐terminal domain of about 120 residues with the fibulins23; in fibrillin‐1, this module may be involved in homotypic interactions.24
Tissue organisation of microfibrils
Fibrillin microfibrils are widely distributed extracellular matrix multimolecular assemblies comprised of fibrillin and other proteins. The microfibrils endow elastic and non‐elastic connective tissues with long range elasticity. They direct tropoelastin deposition during elastic fibrillogenesis and form an outer mantle for mature elastic fibres. Microfibril arrays are also abundant in dynamic tissues that do not express elastin, such as the ciliary zonules of the eye. Analysis of fibrillin‐rich microfibrils by transmission electron microscopy has revealed them to have a diameter of 8–12 nm, a tubular appearance, and beaded periodicity, and to contain glycoproteins. Isolated fibrillin‐rich microfibrils have a complex “beads on a string” appearance, and are extensible.
Microfibrils form loosely packed bundles in roughly parallel alignment. These bundles adopt tissue‐specific architectures that are dictated by cells, and by the strength and direction of forces put upon the tissue. They are found in locations that are subject to repeated mechanical stresses, and in the proximity of basement membranes, and they serve a critical biomechanical anchoring role in dynamic connective tissues.25
Elastic fibre formation is a developmentally regulated process in which tropoelastin (the soluble precursor of mature elastin) is deposited on a preformed template of fibrillin microfibrils.26 Mature elastic fibres are a composite biomaterial with an outer microfibrillar mantle and an inner core of amorphous cross‐linked elastin with some embedded microfibrils. The proportion of microfibrils to elastin appears to decline with age, with adult elastic fibres often having only sparse peripheral mantles of microfibrils.
A recent investigation by mass spectrometry of the composition of purified fibrillin‐rich microfibrils from non‐elastic and elastic tissues showed that in all microfibril preparations, fibrillin‐1 was abundant and the only fibrillin isoform detected. Isolated microfibrils, extracted from tissues by enzyme digestions or homogenisation, have a “beads on a string” appearance with untensioned periodicity of ∼56 nm.27,28 Unextracted hydrated zonular microfibrils appeared, by quick freeze deep etch microscopy, to be more tubular, suggesting that molecular components are lost or that there is a major molecular rearrangement on extraction.29 The molecular basis of the “beads”, and fibrillin‐1 alignment in microfibrils remain unclear.
Models of fibrillin‐1 alignment
The details of the precise molecular architecture of fibrillin‐1 alignment within microfibrils are not entirely clear, and several models have been proposed. An intermolecular fibrillin‐1 transglutaminase cross‐link30 provides molecular constraints, although mass spectrometry has shown that not all fibrillin‐1 molecules within tissue microfibrils are cross‐linked.31
The “hinge” model, based on detailed scanning transmission electron microscopy, mass mapping, automated electron tomography, and atomic force microscopy data,28,32,33,34 predicts maturation from an initial parallel head to tail alignment to an approximately one third stagger (∼100 nm) that would allow transglutaminase cross‐link formation, and further packing into a more energetically favourable ∼56 nm untensioned form.
The one third staggered model was suggested on the basis of extrapolation of molecular dimensions,35 the crystal structure of fibrillin‐1 cbEGF/TB/cbEGF domain arrays,36 and calcium binding studies of TB/cbEGF flexibility.37
Interested readers can find further discussion of issues surrounding fibrillin‐1 alignment and microfibrillar architecture in Baldock et al,28 Davis et al,29, Kielty et al,34 and Lee et al.36
Early studies of isolated microfibrils revealed a number of highly stretched microfibrils with periodicities up to ∼150 nm, which suggested that microfibrils may have elastic properties.27 Stretching of isolated intact microfibrils has been achieved using surface tension forces and molecular combing.33 These studies showed that microfibrils behave as relatively stiff elastic filaments which can perform anchoring roles in ciliary zonules and other basement membrane interfaces. It was confirmed by x ray diffraction that hydrated microfibril bundles are elastic, and suggested that elasticity in microfibril‐rich tissues may arise, in part, from reversible alterations in supra‐microfibrillar arrangements. Proteolytic damage to microfibrils that may occur in MFS or ageing, may generate stretched microfibrils that may have lost their elastic properties.25,38
Assembly of microfibrils
Today, a fragmented picture has emerged of the events and molecules involved in the assembly process from profibrillins to mature tissue microfibrils. The mechanisms include profibrillin processing, self‐assembly, regulatory events, cross‐link formation, and maturation of microfibrils.
Microfibril assembly in individuals with MFS
Early immunofluorescence studies with dermal fibroblasts or skin biopsies obtained from individuals with MFS showed reduced or qualitatively altered fibrillin networks as compared to controls.39,40,41 However, such altered patterns were not observed in all samples.41,42 One possible interpretation of these data is that a subset of mutations in fibrillin‐1 compromises the assembly process and thus the formation of microfibril networks. Further evidence for this interpretation comes from pulse chase experiments using dermal fibroblasts from individuals with MFS. These analyses showed deficiencies at different levels such as fibrillin synthesis and secretion, as well as deposition into the extracellular matrix.43,44 Despite the differences in the secreted amount of fibrillin, a large portion of the fibroblasts studied showed impaired incorporation of the mutant fibrillin into the extracellular matrix, suggesting functional disturbances in early stages of the assembly mechanism.45,46 In cases where higher order assembly into beaded microfibrils was observed, the ultrastructural appearance showed several types of abnormalities including diffuse, frayed, or poorly defined interbead domains, or variable interbead periodicities.47,48,49 These consequences potentially could also emerge from functional problems in the assembly mechanism. In summary, although molecular evidence is still lacking, it seems clear that a certain subset of mutations in fibrillin‐1 leading to MFS and other microfibrillopathies directly affects microfibril assembly mechanisms.
Role of propeptide processing in fibrillin assembly
Fibrillins are phylogenetically old proteins occurring in species from jellyfish to human. All known fibrillins possess highly conserved basic recognition sites (RX(K/R)R) for processing by endoproteinases of the furin/PACE type within the unique N‐ and C‐terminal domains. It has been shown by several groups and methodologies that fibrillin‐1 is indeed processed at these recognition sites.22,50,51,52,53,54 Processing produces a 17 or 20 residue N‐terminal propeptide depending on the actual cleavage site for the signal peptide, and a 140 residue C‐terminal propeptide. Due to the size of the propeptide, C‐terminal processing is accessed much better experimentally and thus has received more attention. For fibrillin‐1, it has been demonstrated that processing of the C‐terminal propeptide is required for deposition into the extracellular matrix, suggesting that profibrillin‐1 conversion to mature fibrillin‐1 plays a regulatory role in assembly into higher order aggregates.43,50,53 On the molecular level, it is not clear how the presence of a C‐terminal propeptide prevents matrix deposition and assembly. The propeptide may interact and mask self‐assembly sites in fibrillin‐1.24 A mutation in fibrillin‐1 (R2726W) associated with isolated skeletal features of MFS was shown to interfere with normal processing at the C‐terminal end and thus disturbed the incorporation of the mutated protein into the extracellular matrix.50 Other mutations close to processing sites may have similar consequences. Virtually no information is available as regards the functional role of the N‐terminal processing. However, by analogy, it is predicted that the N‐terminal propeptide also regulates assembly of fibrillins.
Self‐assembly of fibrillins
As described in the “Tissue organisation of microfibrils” section, the molecular organisation of fibrillin‐1 in microfibrils has been analysed by various groups resulting in a number of different models for the alignment of fibrillin in microfibrils.7,28,30,35,36,51 Despite the differences in these models in terms of stagger and molecular condensation of individual molecules, common to all models is a head to tail orientation of fibrillin‐1 molecules as originally proposed by Sakai and coworkers in 1991.7 Another commonly accepted property of microfibrils is the involvement of six to eight fibrillin molecules per cross‐section of the interbead region.28,55,56
Correlation of antibody epitopes in the fibrillin‐1 molecule with the location of the corresponding epitopes in microfibrils clearly revealed that the terminal ends of the fibrillin molecules are situated in or close to the beads.7,28,51 Data obtained with recombinant fibrillin‐1 fragments have established direct interaction in a homotypic N‐ to C‐terminal fashion.57 These results established a linear head to tail self‐assembly mechanism for fibrillin‐1. Heterotypic interactions between fibrillin‐1 and fibrillin‐2 in an N‐ to C‐terminal fashion suggested that both fibrillin isoforms can be organised within the same microfibril,57 and colocalisation of both isoforms has indeed been demonstrated by double immunogold labelling in tissue microfibrils.8 Electron microscopy after rotary shadowing of full length recombinant fibrillin‐1 suggested that the interaction epitopes are relatively close to the terminal ends.57 These results were further substantiated by analyses of smaller overlapping fibrillin‐1 fragments in various ligand interaction assays, positioning the interaction sites in the N‐terminal region encoded by exons 1–8 and the C‐terminal region encoded by exons 57–65.24
In addition to a linear head to tail self‐interaction, there is evidence that lateral homotypic interactions also play a role in fibrillin assembly. Reducible homodimer formation early during biosynthesis was observed for recombinant fragments of fibrillin‐1 and ‐2 spanning from the proline and glycine‐rich domains, respectively, to the second 8‐Cys/TB domain,58 as well as for smaller recombinant fragments of the proline‐rich region of fibrillin‐1 and the glycine‐rich region of fibrillin‐2 including flanking domains.59 Additionally, homotypic lateral interactions have been observed with an N‐terminal recombinant fibrillin‐1 fragment encoded by exons 1–8, as well as with an C‐terminal fragment encoded by exons 57–65.24 The lateral homotypic interactions between N‐terminally and C‐terminally located fragments as well as the head to tail linear interactions between the N‐ and C‐terminal ends are of high affinity with dissociation constants in the low nanomolar range, indicating that both types of mechanisms are highly relevant for initial assembly stages.24,57
Intermolecular cross‐link formation in fibrillin assembly
Two types of intermolecular cross‐links important for the stability of microfibrils have been identified: reducible disulfide bonds and non‐reducible ε(γ‐glutamyl)lysine cross‐links. Intermolecular disulfide bond formation is apparent early in the assembly of microfibrils since higher molecular weight disulfide‐bonded aggregates containing fibrillin can be observed after a few hours in cell or organ cultures.22,60 Most of the highly conserved cysteine residues in fibrillins are predicted to stabilise individual domains through intramolecular disulfide bonds.17,35,36 One cysteine residue in the first hybrid domain of human fibrillin‐1 and ‐2 has been shown to be available for intermolecular cross‐links on the surface of the molecule.22
Additional data suggest that other cysteine residues, which are normally involved in intramolecular domain stabilisation, may be reshuffled to participate in intermolecular cross‐links. Cysteine residues in the first and second 8‐Cys/TB domain have been suggested for such a role based on the propensity of various recombinant fibrillin‐1 and ‐2 fragments to form reducible homodimers.58,59 Such a mechanism would likely require the enzymatic activity of one or more protein disulfide isomerases on the cell surface or in the extracellular matrix.
Non‐reducible ε(γ‐glutamyl)lysine cross‐links, catalysed by transglutaminases, have been identified in microfibrils extracted from various tissues.30,61,62 Detailed analyses of microfibrils from human tissues have identified transglutaminase cross‐links in the N‐ and C‐terminal regions of fibrillin‐1, as well as a high overall content of these cross‐links.30 In addition to fibrillin‐1, another prominent protein in microfibrils, microfibril‐associated glycoprotein‐1 (MAGP‐1), was also characterised as a substrate for transglutaminase.63 It is possible that besides homotypic fibrillin‐1 transglutaminase cross‐links, heterotypic fibrillin‐1‐MAGP‐1 cross‐links may be present in microfibrils. Zonular fibres in the eye have been demonstrated to be a target for transglutaminase 2.64 Biomechanical analyses of microfibrils suggested that transglutaminase cross‐links play an important role in strengthening the microfibrils.65 Other potential roles of the transglutaminase cross‐links may include correct lateral alignment of fibrillin or other molecules as a prerequisite for downstream assembly events. Mutations disrupting transglutaminase cross‐link sites likely result in serious consequences for microfibril assembly and stability. To gain insight into such potential mechanisms in MFS and other microfibrillopathies, it will be important to identify the exact amino acid residues involved and the time course of transglutaminase cross‐link formation.
Accessory molecules important for fibrillin assembly
In addition to self‐assembly and cross‐linking mechanisms, other molecules may be involved in the assembly process of microfibrils. Several regions in fibrillin‐1 have been identified as interacting with heparin/heparan sulfate with high affinity.66,67,68 In cell culture assembly assays, these glycosaminoglycans inhibit the formation of microfibrillar networks,66,67 leading to the hypothesis that heparan sulfate or heparan sulfate containing proteoglycans may have regulatory functions in the assembly of microfibrils. Recently, the heparan sulfate containing proteoglycan perlecan was identified as a molecule which interacts with fibrillin‐1 and with microfibrils close to basement membrane zones.69 Reduced amounts of microfibrils in basement membrane zones of perlecan null mice may reflect a potential role for perlecan in microfibril assembly.69
Microfibril‐associated proteins
In addition to fibrillin, several other proteins are integral components of the microfibrils or associated with them. These can be grouped into small non‐fibrillin proteins that are integral parts of fibrillin‐rich microfibrils (table 1) and proteins that can associate with fibrillin‐rich microfibrils but do not serve an integral structural function (table 2). We will not attempt a comprehensive review of the non‐fibrillin proteins but rather will summarise the most important functions that have been attributed to these proteins and their interaction with fibrillin. Readers are referred to Gibson70 and the references in the tables 1 and 2 for further information. It appears likely that more interacting proteins remain to be discovered in light of a recent proteomics study of fibrillin‐rich microfibrils showing copurification of a number of proteins in addition to fibrillin‐1. For instance, γ‐crystallin copurified with zonular microfibrils, suggesting an interaction that could contribute to zonule anchorage to the lens.31
Table 1 Small non‐fibrillin proteins that are integral parts of fibrillin‐rich microfibrils in at least some tissues and developmental stages.
| Protein | Chromosome | Potential functions |
|---|---|---|
| MAGP‐1 | 1p36.1–p35 | Tropoelastin deposition78 |
| Tropoelastin binding76,89 | ||
| Binding to fibrillin‐176,90 | ||
| Binding to fibrillin‐291 | ||
| Ternary complex with | ||
| tropoelastin and biglycan77 | ||
| Posttranslational modifications92 | ||
| Ternary complex with | ||
| tropoelastin and decorin93 | ||
| Binding to type VI collagen79 | ||
| Substrate for transglutaminase63 | ||
| MAGP‐2 | 12p12.3–p13.1 | Binding to fibrillin‐174,90 |
| Binding to fibrillin‐274 | ||
| RGD‐mediated cell attachment94 | ||
| Interaction with Jagged180 | ||
| AAAP‐40 | 5q32–q33.2 | 40 kDa protein95,96 |
| (MAGP‐3) | ||
| MFAP1 (AMP) | 15q15–q21 | 54 kDa protein that is processed |
| to a 36 kDa protein97,98 | ||
| MFAP3 | 5q32–q33.1 | 41 kDa serine‐rich protein99 |
| MFAP4 | 17p11.2 | Colocalisation to elastic fibres100 |
| (MAGP‐36) | Role in elastogenesis101 |
Table 2 Proteins that can associate with fibrillin‐rich microfibrils but do not serve an integral structural function in them.
| Protein | Class of protein | Potential biological role of interaction |
|---|---|---|
| Elastin | – | Tropoelastin deposition78 |
| Regulation of microfibril formation75 | ||
| LTBP‐1 | Fibrillin‐LTBP | Sequestering of latent TGFβ102 |
| LTBP‐2 | Fibrillin‐LTBP | Structural role?103 |
| LTBP‐4 | Fibrillin‐LTBP | Sequestering of latent TGFβ102 |
| Versican | Proteoglycan (hyalectin) | Link fibrillin‐microfibrils to versican/hyaluronan network104 |
| Perlecan | Proteoglycan (hyalectin) | Anchoring microfibrils to basement membranes and in the |
| biogenesis of microfibrils69 | ||
| Heparin/heparan sulfate | Proteoglycan | Binding of related heparan sulfate chains may regulate cell‐surface |
| assembly of fibrillin66 | ||
| Decorin | Small dermatan sulfate proteoglycan | Induction of fibrillin‐1 expression in renal fibroblasts and mesangial cells105 |
| Biglycan | Small dermatan sulfate proteoglycan | Role in elastogenesis77 |
| Fibulin‐2 | Fibulin | Mediate/modulate attachment of fibrillin to tropoelastin106 |
| Fibulin‐5 | Fibulin | Regulation of the initial deposition of tropoelastin on to microfibrils107 |
| BMP‐7 | Bone morphogenetic protein | Regulation of limb patterning12 |
| EMILIN‐1 | Elastin‐microfibril interface located proteins | Role in elastinogenesis108 |
Structure and assembly of microfibrils
One role of non‐fibrillin microfibrillar proteins is as structural constituents of the microfibrils. For instance, microfibril‐associated glycoprotein‐1 (MAGP‐1) is a small glycoprotein with an apparent molecular weight of 31 kDa that is covalently bound to microfibrils by disulfide linkages71 and is specifically located on the beads of the beaded‐filament structure of the microfibrils.72 MAGP‐2 has a more restricted tissue distribution than MAGP‐1,73 suggesting it may have tissue‐ or developmental stage‐specific functions. MAGP‐1 and MAGP‐2 bind to distinct regions of fibrillin‐1 and it has been suggested that this may help to regulate microfibrillar assembly.74
Interaction with other matrix components
Interactions between fibrillin‐rich microfibrils and tropoelastin are important for the formation of elastic fibres. In addition to interactions between fibrillin‐1 and tropoelastin,75 MAGP‐1 interacts with tropoelastin76,77 in a way that is important for tropoelastin deposition.78 Microfibrils interact with a variety of other extracellular matrix structures and some of these interactions are mediated by non‐fibrillin proteins. For instance, MAGP‐1 binds to the α3 chain of type VI collagen. Since type VI collagen microfibrils and fibrillin‐rich microfibrils are often found near to one another in some extracellular matrices, it is plausible that MAGP‐1 may mediate a molecular interaction between type VI collagen microfibrils and fibrillin‐containing microfibrils.79
Interaction with cells
It has become increasingly clear that fibrillin‐rich microfibrils have functions that are not directly related to structural integrity but rather have to do with growth factor metabolism and triggering cellular signals. As will be discussed in more detail below, the latent transforming growth factor‐β binding proteins (LTBP) are a family of secreted glycoproteins, three of which play an important role in the regulation of TGFβ regulation.15 In addition to TGFβ, microfibrils could conceivably be involved in the regulation of other growth factors, although little experimental evidence is available at this point.14 MAGP‐2 can interact with Jagged1 and induce its shedding; it is thus conceivable that MAGP‐2 may be able to modulate the Notch signalling pathway.80 There is evidence of interaction between fibrillin‐1 and at least one bone morphogenetic protein (BMP),81,82 and fibrillin‐2‐rich microfibrils and BMP‐7 have been shown to functionally interact in the regulation of limb patterning in a mouse model.12 Finally, an interesting area of research is the question whether novel cell signalling pathways are triggered in MFS or other diseases of the microfibrils, and whether fragments of the proteins of the microfibrils can acquire novel signalling properties through exposure of otherwise cryptic binding sites, a phenomenon that has been well described in several other disorders such as osteoarthritis.83 As will be discussed below, there is evidence that fibrillin fragments can induce matrix metalloproteinase (MMP) expression, and it is well known that elastin fragments can induce MMP expression.84,85
Enzymatic activity
The enzyme lysyl oxidase, which is involved in the cross‐linking of tropoelastin monomers, has been localised to the interface between extracellular bundles of amorphous elastin and the microfibrils.86 Although there is no evidence of a direct interaction between fibrillin and lysyl oxidase to date, it is plausible that interactions with fibrillin or other microfibrillar components might be important for elastogenesis.70 Interestingly, ADAMTS10,87 an extracellular matrix protease, is mutated in the autosomal recessive form of Weill‐Marchesani syndrome (WMS).88 The dominant form of WMS is caused by mutations in fibrillin‐1, suggesting, perhaps, a potential interaction between ADAMTS10 and fibrillin‐1. However, the substrates of ADAMTS10 or indeed other potential functions of this molecule remain to be elucidated.
Mutations in FBN1, FBN2, and other genes associated with MFS and related phenotypes
Mutations in FBN1 were discovered in individuals with MFS in 1991,109 and subsequently mutations in FBN2 were discovered in individuals with a phenotypically related disorder, congenital contractural arachnodactyly (CCA).110 More recently, mutations in the genes for TGFBR1 and TGFBR2 were found in several disorders with varying degrees of overlap with classic MFS.111,112,113 Genetic loci for other forms of isolated aortic dilatation and dissection have been identified. The identification of mutations in these genes has provided significant insight into the pathogenetic pathways involved in MFS and related disorders, and further insight is to be expected from characterising the full spectrum of mutations associated with these disorders and from identifying the full set of genes in which mutations cause related disorders of connective tissue. The following sections present the current state of knowledge on FBN1, FBN2, and TGFBR2 mutations and on loci involved in isolated aortic aneurysm and dissection.
Mutation analysis of the FBN1 gene in individuals with MFS: sensitivity, methods, and clinical indications
In most instances, the diagnosis of MFS can be made on clinical grounds. A set of clinical diagnostic criteria, as summarised in the Ghent nosology,114 define major criteria with high diagnostic specificity and minor criteria with less specificity. In order to make a diagnosis of MFS, the Ghent nosology requires a combination of major criteria in at least two organ systems and involvement of a third organ system. In the majority of cases, these criteria allow the establishment or exclusion of the diagnosis. However, the interpretation of these criteria is not always obvious for a number of reasons. Firstly, MFS is known for its extensive phenotypic variability both within and between families, which may cause underdiagnosis of the condition.115 Secondly, establishing a diagnosis of MFS in children can be difficult because several manifestations of MFS are age‐dependent and may not yet be present in childhood. Thirdly, clinical overlap exists between MFS and other, so called Marfan‐like conditions which share some of the features of MFS but do not necessarily have the same outcome.
Therefore, if the Ghent criteria are fulfilled, then the diagnosis of MFS is certain. In adult patients presenting with a small number of non‐specific skeletal manifestations often seen in MFS, the diagnosis is unlikely, but a full clinical evaluation including echocardiography and ophthalmologic examination is indicated.
In situations of clinical uncertainty, molecular analysis of the FBN1 gene therefore seems a logical aid to the clinical diagnosis of MFS. In practice, however, the large and complex structure of the FBN1 gene and the wide scope of FBN1 mutations have hampered clinical implementation of FBN1 testing. Moreover, literature data show great variation in detection rates of FBN1 mutations and methodologies used. This can be accounted for by a variety of factors such as the type of mutational analysis method, the substrate (genomic versus cDNA) and, most importantly, the accuracy of the clinical diagnosis. The studies that have looked at the sensitivity and specificity of FBN1 mutation analysis show substantial differences in one or several of these factors.
The first mutational studies, performed on cDNA, showed low efficiency of FBN1 mutation detection using SSCP (single stranded conformation polymorphism) and yielded mutation rates of 9% to 23%.116,117 Subsequent studies, using genomic DNA as template for analysis of the 65 individual exons of the FBN1 gene, obtained higher detection rates but varied according to the type of screening method used.118,119,120 Initially the best results were obtained with CSGE (conformation sensitive gel electrophoresis), with detection rates ranging from 57% to 90%.42,121,122 Subsequent studies have shown that mutation detection by DHPLC (denaturing high performance liquid chromatography) is highly efficient,123 although a relatively high false‐positive rate may be an issue.124 It now appears that DHPLC is the most efficient approach for mutation detection also because of its potential for automation when combined with robotic PCR.
The most important factor influencing the mutation detection rate appears to be the clinical homogeneity or heterogeneity of the patient population. Indeed, several studies have shown that the incidence of FBN1 mutations is significantly higher in patients who fulfil the MFS diagnostic criteria than in patients who do not. In a large study including a cohort of 94 MFS patients and 77 patients with MFS related phenotypes, Loeys et al125 found FBN1 detection rates of 66% versus 5%, respectively. They demonstrated that fulfilling the clinical diagnosis of MFS in itself is a good predictor of the outcome of FBN1 mutation analysis. Katzke et al120 supported these findings by their study which showed a much higher incidence of mutations in a group of MFS patients versus those with an MFS related condition, and concluded that clinical overdiagnosis is the most important explanation for low FBN1 mutation detection rates. Two other studies123,126 also showed that the majority of patients in whom an FBN1 mutation was found, met the clinical diagnosis of MFS, supporting the opinion that the robustness of selection criteria is the most important determinant of the outcome of mutational studies.
Several clinical situations can occur in which molecular studies of the FBN1 gene may be helpful. In patients who present with skeletal, cardiovascular, and/or possibly other manifestations of MFS but have no involvement of the ocular system, it can be difficult to establish the diagnosis strictly on clinical grounds, particularly in the absence of a positive family history. Here, however, MRI studies can reveal the presence of dural ectasia in which case the diagnostic criteria may still be met. Several mutational studies which report FBN1 mutations in Marfan‐like patients have not verified the presence or absence of dural ectasia, so that the possibility remains that the diagnostic criteria are in fact met. In cases where no definitive conclusion can be reached with the clinical data, molecular analysis of the FBN1 gene is an alternative option.
Because of the evolving nature of the phenotype, particularly so for the cardiovascular and skeletal manifestations, children with suspected MFS may not yet fulfil the diagnostic criteria. In those instances, it is better to postpone a final diagnosis until later. The identification of an FBN1 mutation in children or young adults not (yet) fulfilling the diagnostic criteria can help to identify those who need to be clinically followed with particular attention.
FBN1 mutations have been identified in a range of phenotypes, the type 1 fibrillinopathies, with greater or lesser degrees of clinical overlap with MFS (table 3). A decision as to whether mutation analysis is indicated when such phenotypes are suspected needs to be made on an individual basis.
Table 3 Type 1 fibrillinopathies.
| Syndrome | Clinical features | Reference |
|---|---|---|
| MFS | See text | See text |
| Neonatal MFS | Severe end of clinical spectrum | Kainulainen et al,117 Booms et al129 |
| Atypically severe MFS | Severe and early onset cardiovascular complications | Putnam et al,130 Tiecke et al,131 |
| Ectopia lentis | Mainly ocular findings | Lönnqvist et al,132 Ades et al,133 |
| Kyphoscoliosis | Progressive kyphoscoliosis of variable severity | Ades et al134 |
| Familial arachnodactyly | Dolichostenomelia and arachnodactyly | Hayward et al135 |
| Familial thoracic ascending aortic | See text | |
| aneurysms and dissections | ||
| MASS phenotype | Mitral valve prolapse, aortic dilatation without dissection, | Dietz et al136 |
| skeletal and skin abnormalities | ||
| Shprintzen‐Goldberg syndrome | Craniosynostosis, a marfanoid habitus, and skeletal, | Sood et al,137 Kosaki et al138 Robinson et al139 |
| neurological, cardiovascular, and connective tissue anomalies | ||
| Isolated skeletal features | Tall stature, scoliosis, pectus excavatum, arachnodactyly | Milewicz et al50 |
| New variant of MFS | Skeletal features of MFS, joint contractures, ectopia lentis, | Ståhl‐Hallengren et al,140 Black et al,141 |
| no cardiovascular manifestations | ||
| Weill‐Marchesani syndrome | Short stature, brachydactyly, joint stiffness, and | Faivre et al142 |
| (autosomal dominant) | characteristic eye abnormalities |
Although classic MFS is by far the most common disorder associated with FBN1 mutations, several other disorders with overlapping clinical findings have been described due to mutations in FBN1.
In addition to the more or less well delineated disorders listed in table 3, FBN1 mutations can be found in individuals with Marfan‐like disorders who do not fulfil the criteria of the Ghent nosology.127 It is recommend that accepted clinical guidelines for the care and management of MFS are applied in these cases even if the criteria of the Ghent nosology are not fulfilled, because it is possible that complications such as aortic dilatation can emerge at any age.
Finally, the availability of a molecular test also allows prenatal or preimplantation diagnosis for prospective parents. This is one option for which requests appear to be steadily increasing.
In summary, after stringent clinical selection, a detection rate of up to 90% is currently achievable in FBN1 molecular testing of patients with classic MFS.128 This allows the implementation of mutational studies in clinical practice.
The UMD FBN1 database: a database for FBN1 mutations implicated in MFS and related phenotypes
The UMD FBN1 database (http://www.umd.be) was created in 1995 in an effort to standardise the information regarding FBN1 mutations using UMD (Universal Mutation Database) software.143,144,145,146,147,148,149 The database follows the guidelines on mutation databases of the Hugo Mutation Database Initiative including nomenclature of mutations.150
The mutation records of the database include point mutations, large and small deletions, insertions, and splice mutations in the FBN1 gene. Each record contains the molecular and clinical data for a given mutation in a standardised, easily accessible, and summary form; if available, data on fibrillin protein biosynthesis classification groups45 are included.
To date, 601 FBN1 mutations are available online. The mutations are spread throughout almost the entire gene without obvious predilection for any given region. Approximately 12% of mutations are recurrent.149
The mutation studies performed to date generally have concentrated on screening the 65 coding exons of FBN1. For the most part, methods capable of detecting larger deletions were not applied and it is not clear how much flanking sequence or other regions of the gene were investigated. It is also unclear whether, once a mutation was identified, the remaining gene regions were regularly and fully evaluated.
Many different kinds of mutation have been identified in FBN1. Point mutations are the most common mutational event, with nonsense and missense mutations comprising about 10% and 60% of all reported mutations. The most common missense mutations substitute cysteine residues that form disulfide bonds within one of the cbEGF or 8‐Cys domains, but missense mutations creating novel cysteine residues in these modules are also common. The majority of the remaining mutations in these modules affect residues of the calcium consensus sequence. About a quarter of missense mutations affect modules other than cbEGF, and for the most part, the pathophysiological mechanisms of these mutations remain unclear.
Small insertions, deletions, or duplications represent about 13% of all reported mutations. The majority of these mutations create a premature termination codon (PTC). Another 13% of reported mutations consist of various classes of splicing errors, most commonly affecting canonical splice sequences at exon/intron boundaries. Many splice site mutations in FBN1 result in in‐frame exon skipping, such that the mutant fibrillin‐1 lacks an entire cbEGF domain. Such mutations can be associated with a particularly severe phenotype.151 Some exon‐skipping mutations in FBN1 result in a frameshift152 with reduced mutant RNA levels through nonsense‐mediated decay of the mutant transcript.153 A nonsense mutation136 and a silent exonic mutation154 in exon 51 have been reported as inducing in‐frame skipping of the entire exon 51 and demonstrate the existence of an exonic splicing enhancer.155,156
Global analysis of FBN1 mutations reveals two classes of mutations. The first type, which represents more than one third of the mutations, contains mutations predicted to result in shortened fibrillin‐1 molecules, including nonsense mutations, splicing errors, insertions, and duplications, as well as in‐frame or out‐of‐frame deletions. These mutations are likely to result in nonsense‐mediated decay resulting in reduction in the level of the mutant allele. The second type represents slightly less than two thirds of the mutations and contains missense mutations, mostly located in cbEGF‐like modules. They can be subclassified into: (a) mutations creating or substituting cysteine residues potentially implicated in disulfide bonding and consequently in the correct folding of the monomer; (b) amino acids implicated in calcium binding and subsequently in interdomain linkage, structural integrity of affected domains, and increased protease susceptibility; and (c) other mutations that might affect the conformation of affected modules, interdomain packing, or other functions such as protein‐protein interactions.
Elucidating the molecular basis of MFS and related fibrillinopathies is the major goal of the teams working on this subject.149,157 The extreme clinical variability, the difficulties associated with clinical diagnosis, and the low detection rate of mutations in this large gene all conspire to negatively impact on progress. At present it is not possible to predict the phenotype for a given FBN1 mutation. On the one hand, mutations affecting different positions within a given module may be associated with quite different phenotypes. On the other hand, mutations affecting an analogous residue within two different modules may also be associated with differing phenotypes. Therefore, it is apparent that neither the location of the affected structural module in the protein nor the position of the altered residue is, in itself, sufficient to predict potential genotype‐phenotype correlations.158 The high degree of intrafamilial variability suggests that environmental and perhaps stochastic factors or modifying genes are important for the phenotypic expression of disease. The level of the expression of the normal fibrillin‐1 allele159 and hyperhomocysteinaemia related to the C677T methylenetetrahydrofolate reductase polymorphism160 have been proposed as factors that modify the clinical severity of MFS. The elucidation of the full range of modifying factors in MFS represents an interesting area for further research.
Mutations in TGFBR1 and TGFBR2
Signalling by TGFβ family cytokines controls a variety of cellular processes including proliferation, differentiation, and apoptosis; propagation of signalling into the cell is mediated by a family of type 1 and type 2 receptors including the type 1 and type 2 TGFβ receptor. TGFβ binds first to type II receptors, allowing subsequent incorporation of type I receptors into a ligand‐receptor complex involving a TGFβ dimer and four receptor molecules. The signal is then propagated into the cell by means of phosphorylation of the Smad proteins.161
Linkage to chromosome 3p24.2–p25 was demonstrated for a large family with a Marfan‐like phenotype for whom linkage to FBN1 and FBN2 had previously been excluded.162,163 This disorder has been termed MFS type II (MIM 154705) and shares some of the cardiovascular and skeletal features of classic MFS.
Identification of a chromosomal breakpoint disrupting the gene encoding the TGFβ receptor 2 (TGFBR2) in a boy with short stature, dural ectasia, and several skeletal and cardiovascular manifestations of MFS led to the identification of three further missense mutations in four families or individuals with manifestations of MFS in whom FBN1 mutations had been ruled out (one mutation was found in two unrelated families).111 All the mutations were found in the serine‐threonine kinase domain of the TGFβ receptor 2.
More recently, a new aortic aneurysm syndrome with hypertelorism, bifid uvula or cleft palate, and generalised arterial tortuosity with ascending aortic aneurysm together with other findings such as craniosynostosis, mental retardation, and congenital heart disease was described; this disorder, Loeys‐Dietz aortic aneurysm syndrome (LDS; MIM 609192), was shown to be associated with mutations in the genes for either TGFβ receptor type 1 or TGFβ receptor type 2 resulting in perturbations of TGFβ signalling.112 As will be noted below, mutations of the arginine at position 460 of TGFBR2 have been identified in individuals with thoracic ascending aortic aneurysms and dissections (TAAD), although there appears to be some degree of phenotypic overlap with LDS in many affected individuals.
Microsatellite instability (MSI) is a prominent feature in hereditary non‐polyposis colorectal cancer (HNPCC) and some forms of acquired colon cancer. Defects in mismatch repair (MMR) genes and associated coding region MSI can cause frameshift mutations with functional inactivation of affected genes, thereby providing a growth advantage to MMR deficient cells. TGFBR2 is one of the most commonly affected genes with mutations in a polyadenine tract in exon 3 being found in up to 90% of cases of microsatellite‐instable sporadic and HNPCC associated colon cancer.164 Germline mutations in TGFBR2, however, are a rare cause of HNPCC.165 Interestingly, one mutation (R528H) was found both as a somatic mutation in colon cancer166,167 and as a germline mutation in LDS.112 At present, there is no evidence that individuals with LDS are at increased risk for colon cancer.
It should be noted that there has been some controversy as to whether certain TGFBR2 mutations lead to a phenotype that is identical or at least very similar to that of classic MFS, thus justifying the diagnosis of type 2 MFS (MFS2). The question about locus heterogeneity for MFS has been addressed by several studies. Historical linkage data in MFS families showed a cumulative LOD score for the FBN1 locus in excess of 100, which provides evidence for a single predominant locus for MFS. On the other hand, none of the individuals with LDS112 fulfilled the clinical diagnostic criteria for MFS. In a previous study, Loeys et al128 identified 86 FBN1 mutations in a cohort of 93 patients with classic MFS. None of the remaining patients had a TGFBR2 or TGFBR1 mutation. Overall, this suggests strongly that FBN1 is the predominant if not sole locus for MFS.128 However, there have been reports of individuals with TGFBR2 mutations diagnosed with MFS2 without features characterising LDS.168 Given that the aneurysms in LDS appear to be more aggressive than those in MFS and the fact that the cardiovascular involvement is also characterised by arterial aneurysms throughout the arterial tree and marked arterial tortuosity, it is clinically very important to be aware of the potential differences between these two syndromes. Detailed clinical characterisation of individuals with TGFBR2 mutations will be required to determine if a subset of these mutations is associated with MFS2 or whether the diagnosis of LDS is more appropriate.
Mutations in FBN2 and congenital contractural arachnodactyly
The discovery of a second fibrillin gene led to the genetic association of fibrillin‐2 encoded by FBN2 with congenital contractural arachnodactyly (CCA). CCA or Beals syndrome is characterised by a marfanoid habitus. In addition to the tall, slender asthenic appearance, most individuals with CCA have crumpled ears, flexion contractures, severe kyphoscoliosis, and muscular hypoplasia.169,170,171,172 The ear abnormalities are characterised as a folded upper helix of the external ear. In most patients, contractions of major joints (knees, elbows, ankles) are present at birth. The proximal interphalangeal joints display flexion contractures (that is, camptodactyly). Contractures of the hip, adducted thumbs, and clubfoot may also occur. Bowed long bones and muscular hypoplasia are additional musculoskeletal findings in CCA. Contractures usually resolve with time. Arachnodactyly (long slender fingers and toes) is present in most individuals with CCA. The greatest morbidity in CCA is caused by progressive kypho/scoliosis that can begin in early infancy. It is present in about half of all affected individuals. The spinal abnormalities are progressive. Severe thoracic cage abnormalities with associated scoliosis may cause restrictive pulmonary disease.173
While CCA shares some clinical characteristics with MFS (table 4), it does not share the usually shortened life expectancy.
Table 4 Clinical features of congenital contractural arachnodactyly.
| Marfanoid habitus |
| Flexion contractures of multiple joints including elbows, knees, hips, fingers |
| Kyphoscoliosis (sometimes severe) |
| Muscular hypoplasia |
| Abnormal pinnae (presenting as crumpled outer helices) |
Putnam et al110 were the first to identify mutations in two unrelated individuals with CCA; both were cysteine substitutions. It is important to note that in contrast to the distribution of FBN1 mutations causing MFS throughout the coding region, the FBN2 mutations so far identified in CCA appear to cluster between exons 23 and 34. The homologous region of FBN1, the so called neonatal region, contains the greatest percentage of mutations from MFS patients at the most severe end of that disorder's clinical spectrum.117 Virtually all of the known FBN2 mutations are of the calcium binding epidermal growth factor‐like (cbEGF) domains.174,175,176,177,178
Molecular studies of only one individual with severe/lethal CCA have been performed.179 This individual had an exon splicing mutation that caused the skipping of exon 34, a cbEGF‐like domain. Significantly, this individual's mother was a somatic mosaic with one third of her fibroblasts also harbouring the same exon 34 mis‐splicing mutation. Therefore, one can speculate that there is a threshold for certain mutations causing skeletal perturbations versus severe developmental abnormalities in the cardiovascular and gastrointestinal systems.
Although individuals with CCA usually do not have aortic involvement, aortic root dilatation does occur in some cases and screening for aortic involvement should be performed in individuals with this disorder.180
Fibrillin‐2 in development and animal models
The temporal and spatial expression of fibrillin‐2 has been examined in several species. In human fetal aorta, antibodies to fibrillin‐2 were found to stain most intensely in the media, where elastic fibres are most abundant. In human elastic cartilage, fibrillin‐2 localised to the cartilaginous core while fibrillin‐1 localised primarily to the surrounding connective tissue.4 Fibrillin‐1 and fibrillin‐2 demonstrate a similar spatial and temporal distribution in most tissues during early human embryonic development. Exceptions included the kidney, liver, rib anlagen, and notochord.181 Similar studies in the developing mouse showed that in most tissues fibrillin‐2 was expressed earlier than fibrillin‐1.182
Studies in the chick have shown that fibrillin‐2 (called JB‐3 in the early literature) is expressed very early in development and is found in the regions of heart development.183 The early expression of the fibrillins has led to speculation that they may mediate the tensile forces that shape the early embryo.184 A possible role for fibrillin‐2 in lung development has been shown in a rat model. Studies of fetal lung explants demonstrated abnormal branch morphogenesis when the explants were incubated with antisense oligonucleotides to fibrillin‐2.185
Browning et al186 described a mouse with syndactyly (sne) that was derived from chemical mutagenesis of murine embryonic stem cells. They showed that sne (now renamed syfp‐3J) was an allele of the sy locus. sy is the shaker‐with‐syndactylism mouse, a radiation mutant with a chromosome 18 (syntenic to human chromosome 5) contiguous gene deletion syndrome.187 The deleted region contains the gene encoding fibrillin‐2. Some spontaneously occurring mouse models with syndactyly also mapped to the sy locus (syfp and syfp‐2J). All three syfp mutations are FBN2 mutations.186,188 Additional evidence that absence of FBN2 leads to syndactyly came from gene targeting studies.12 The FBN2−/− knockout mouse displayed the same type of syndactyly observed in the sy mice. Interestingly, two of the fibrillin‐2 mutations in the syfp mice were outside the neonatal region, that is, the area in which all of the human CCA mutations have been found.188 These findings have led to the obvious speculation that fibrillin‐2 mutations outside the neonatal region, for example, may lead to other human phenotypes.188
Familial thoracic ascending aortic aneurysms and dissections
Cystic medial necrosis (CMN) is known to be associated with syndromes such as MFS, but is more frequently found in the absence of an associated phenotypic syndrome. Reports of families with autosomal dominant inheritance of thoracic aortic aneurysms leading to type A dissections (TAAD) with medial necrosis on pathologic examination indicate that single gene mutations can cause medial necrosis in the absence of an associated syndrome.189,190 Additionally, medial necrosis of the proximal aorta with aneurysms/dissections is associated with other heritable diseases such as Turner syndrome,191 Noonan syndrome,192 Ehlers‐Danlos syndrome,193 patent ductus arteriosus,194,195 and bicuspid aortic valve.196,197
Initial studies showed that first degree relatives of probands with non‐syndromic TAAD have a higher risk of thoracic aortic aneurysms and sudden death compared with a control group.198,199 In addition, these studies support the hypothesis that genetic factors play a role in the aetiology of TAADs in patients who do not have an identified syndrome causing aortic disease. Milewicz and colleagues described six families with aortic aneurysms and dissections, all of whom demonstrated autosomal dominant inheritance associated with decreased penetrance and variable age of onset of the aortic disease.189 Various studies indicate that the aortic disease in the majority of these families is not due to a mutation in the FBN1 gene or other genes encoding vascular proteins, such as COL3A1.189,200,201
More recently, several loci for non‐syndromic TAAD have been mapped. The first locus, termed TAAD1, was mapped to chromosome 5q13–14 with a maximum LOD score of 4.74 with the marker D5S2029.202 This locus was confirmed by an independent study in a Finnish population where approximately one half of the families studied show evidence of linkage to TAAD1.203 The critical interval containing the defective gene maps to a 7.8 cM region. Another locus for familial aortic aneurysms and dissections has been mapped to the long arm of chromosome 11 (11q23–24) using a single large family.204 In contrast to the families linked to TAAD1, the clinical phenotype of the family linked to the FAA1 locus indicated a diffuse vascular aetiology. Apart from dilatation in the sinuses of Valsalva, involvement of other aortic segments and arteries was also observed, such as dilatation in the abdominal aorta and left subclavian artery. In addition, the disease was fully penetrant with aortic imaging in the family described. The FAA1 locus is a rare cause of the vascular condition as indicated by the fact that no other families demonstrate linkage of the phenotype to markers at this locus.
Another locus for TAAD was mapped to a 25 cM region on chromosome 3p24–25 using another large family with multiple members with aneurysms and dissections of the thoracic aorta. The disease in the family was characterised as autosomal dominant with decreased penetrance and variable age of onset.205 Eighteen TAAD families described previously failed to show linkage to 3p24–25, indicating that TAAD2 is a minor locus for TAAD.202,204 It was recently determined that mutations in the transforming growth factor beta receptor type II gene (TGFBR2) is the cause of disease at the TAAD2 locus.113 The TGFBR2 gene was screened for missense, nonsense, and exon splicing errors and mutations were found in four out of 80 unrelated families with familial TAAD, indicating that TGFBR2 mutations are a relatively rare cause of familial TAAD. Although most vascular disease in these families involved ascending aortic aneurysms leading to type A dissections, affected family members also had descending aortic disease and aneurysms of other arteries, including cerebral, carotid, and popliteal aneurysms. Strikingly, all four families carried mutations that affected arginine at amino acid 460 in the intracellular domain, suggesting a mutation hot spot for familial TAAD and establishing a strong genotype‐phenotype correlation between familial TAAD and mutations at this location. Structural analysis of the TGFBR2 serine/threonine kinase domain reveals that R460 is strategically located within a highly conserved region of this domain and that the amino acid substitutions resulting from these mutations will interfere with the receptor's ability to transduce signals. A surprising observation in these families was that there is no evidence of an increased susceptibility to cancer in families with germline TGFBR2 mutations, despite evidence in the literature that somatic TGFBR2 mutations occur in a variety of cancers.
In summary, TAAD is a genetically heterogeneous disease that may be inherited in conjunction with a syndrome or as a non‐syndromic predisposition for TAAD. Studies of familial TAAD due to TGFBR2 mutations have highlighted the dysregulation of the TGFβ pathway as a mechanism leading to aneurysm formation. Clinically, it is important to perform cardiovascular evaluation on first degree relatives of individuals with suspected non‐syndromic familial aortic dissection, because of the possibility of potentially life‐threatening vascular disease in an individual with obvious phenotypic signs of the disorder.
The effects of fibrillin mutations on the structure of fibrillin‐1 domains
In order to understand the effects of fibrillin mutations on the structure and function of the tissues affected by MFS, it will ultimately be necessary to understand the effects of mutations on protein structure. It has not been possible to determine the structure of the entire fibrillin‐1 protein or even of larger fragments thereof. However, studies on recombinant polypeptides comprising up to several cbEGF or 8‐Cys modules have provided significant insight.
The two predominant structural modules in fibrillin‐1 are the calcium‐binding epidermal growth factor‐like domain (cbEGF) and the transforming growth factor β binding protein‐like (TB or 8‐Cys) domain. High resolution structures of the fibrillin‐1 domain fragments cbEGF32–33, TB6, and cbEGF12–13 have been solved previously using nuclear magnetic resonance (NMR) which identifies the solution structure of the molecule.17,35,206 More recently, x ray crystallography has been used to solve the structure of the triple domain fragment, cbEGF22‐TB4‐cbEGF2336 and demonstrated a calcium‐stabilised tetragonal pyramidal conformation. An RGD integrin binding site localises to the tip of a β‐sheet within TB4 and is thus accessible to cell‐surface integrins. Comparative sequence alignments of the linker regions from cbEGF‐TB domains within fibrillin‐1 suggest that the relative orientation of cbEGF22‐TB4 is likely to be preserved at homologous sites within fibrillin‐1. In contrast, the variation in amino acid number and composition of TB‐cbEGF linker sequences suggests that these pairs will adopt different orientations with respect to one another within fibrillin‐1, and may contribute to the biomechanical properties of microfibrils. This is supported by the variability of Kd values of TB‐cbEGF pairs from fibrillin‐1 (see below). A model of a large region of fibrillin‐1 (cbEGF11‐TB5) has been generated from this combined structural information, which suggests that although the protein is in an extended conformation, it is not simply linear. A significant bend is introduced by the packing of cbEGF22 against TB4. Based on this structure, a staggered model for assembly of fibrillin‐1 into the microfibril has been suggested as an alternative to the proposed organisation based on electron microscopic studies.36
Single cbEGF domains expressed from fibrillin‐1 and other proteins usually display low affinity binding in the mM range. However, in fibrillin‐1, and in many other proteins, the cbEGF domains are often arranged as repeating tandem arrays. On covalent linkage of an N‐terminal cbEGF, the affinity of the C‐terminal cbEGF increases.207 The bound calcium, together with the hydrophobic packing interaction, performs a key structural role in restricting interdomain flexibility35,208 and therefore protects the modules against proteolytic cleavage.209,210 Dynamics studies show that the most stable region of a cbEGF pair is in the vicinity of the interdomain calcium‐binding site.208 Analysis of different cbEGF domain pairs has, however, identified a range of affinities from 350 μM to 300 nM, suggesting that primary sequence variation, in addition to the pairwise domain interaction, must also influence affinity.211,212 A study of heterologous TB‐cbEGF domain pairs37 has shown that most of these domain pairs bind Ca2+ considerably more tightly than previously observed, with Kd values as low as 9 nM. These data suggest that under physiological conditions, many fibrillin‐1 cbEGF domains will be fully saturated and may impart rigidity to the native protein. However, the TB6‐cbEGF32 domain pair, with a Kd of 1.6 mM,213 appears the most likely of the TB‐cbEGF domain pairs to be flexible and may contribute to the extensibility and elasticity of the microfibrils.
Insights into the role of a number of disease‐causing FBN1 mutations in the pathogenesis of MFS have been gained from NMR, calcium chelation, and limited proteolysis studies of recombinantly expressed fragments of fibrillin‐1. Reduction of calcium binding caused by substitution of a calcium ligand or destabilisation of the interdomain interface would be predicted to produce a less extended, more flexible structure within a region of fibrillin‐1. This may result in increased proteolytic susceptibility due to exposure of enzyme‐specific cryptic cleavage sites. The effect of a missense mutation on protease susceptibility of a cbEGF domain can, however, be influenced by a number of factors such as the particular residue mutated and the position of the mutant domain within the fibrillin‐1 peptide.
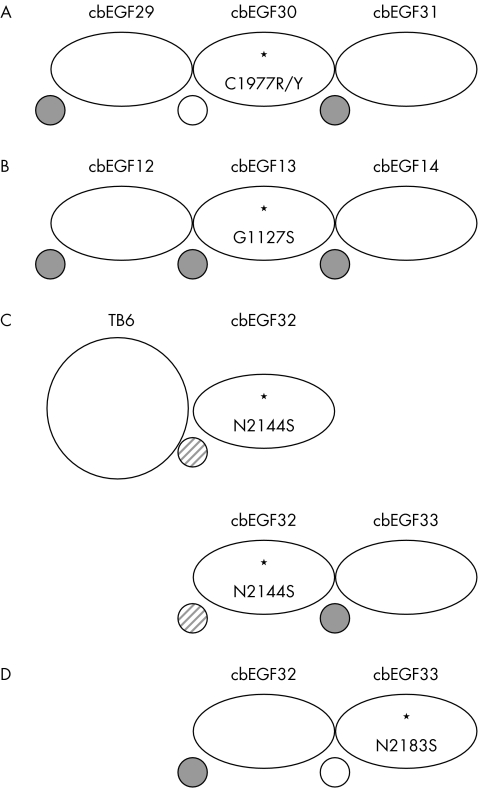
The structural effects of the pathogenic mutations C1977Y and C1977R which disrupt the 1–3 disulphide bond of cbEGF30 and are therefore predicted to cause misfolding, have been studied in a cbEGF29–31 triple construct using the combined methods of NMR, chelation, and limited proteolysis.212 The substitutions caused loss of Ca2+ binding to cbEGF30, consistent with intradomain misfolding and disrupted cbEGF29–30 domain‐domain packing. Surprisingly, the calcium binding properties of cbEGF29 and cbEGF31 were unaffected, suggesting these cysteine substitutions have relatively localised effects confined to the N‐terminal end of the mutant domain (fig 3A). However, a disruption of the 5–6 disulphide bond by a C750G substitution which affects the C‐5 residue of cbEGF7 in an EGF4‐TB3 fragment caused increased proteolytic susceptibility of cbEGF8.214 This is presumably due to disruption of domain packing between cbEGF7 and 8 and hence reduction of the calcium binding affinity of cbEGF8. Cysteine substitutions are therefore likely to have different structural effects, which depend on the particular disulphide bond affected, and hence result in a variety of pathogenic mechanisms.
Figure 3 Schematic illustration of the variable effects of missense mutations on calcium binding in multi‐domain fragments from fibrillin‐1. Calcium binding properties were assessed by NMR, limited proteolysis, and (for A) calcium chelation. Bound calcium is depicted as a grey circle and the absence of detectable binding as an open circle. The grey diagonal striped circle indicates reduced binding.
These studies, together with earlier reports, emphasise the structural heterogeneity that can be introduced into fibrillin‐1 by different FBN1 mutations. In the case of the folding substitution, G1127S, in cbEGF13, it was shown215,216 that the mutant domain retained the ability to bind calcium (fig 3B). Studies210,213 of the effects of the calcium binding substitution, N2144S, in domain pairs demonstrated that, while the structure of the mutant domain was unaffected, its ability to bind Ca2+ was reduced (fig 3C). Calcium binding substitutions which occur in the context of a cbEGF domain pair can, however, result in more significant structural changes.210,217 For example, the protein engineered N2183S substitution in cbEGF33 (fig 3D) resulted in an increased proteolytic susceptibility of the cbEGF32–33 domain pair and the lack of calcium dependent protection indicated the absence of Ca2+ binding to the mutant domain.210 Thus, as with cysteine substitutions, calcium‐binding substitutions may cause variable intramolecular effects dependent upon domain context.
It is evident that the structural effects of different FBN1 missense mutations are complex. In the neonatal region of fibrillin‐1, for example, missense mutations which affect structurally analogous calcium ligands in different cbEGF domains or cause substitution of different ligands coordinating the same Ca2+ (D1113G and N1131Y) produce varying phenotypes. Three missense mutations, K1043R, I1048T, and V1128I, which have no clear structural effect (although the I1048T substitution does introduce a glycosylation consensus sequence) are found to cluster on one face of a model constructed for the cbEGF11–15 region of fibrillin‐1. An unstructured, extended loop, present in cbEGF12 between cysteines 5 and 6, may also localise to this face of the model and be involved in intra‐ or intermolecular contacts.206 Analysis of the model shows that substitutions that may affect the calcium‐binding properties of cbEGF12 give rise to severe phenotypes. An increase in the intrinsic flexibility of this region resulting from defective calcium binding could distort a potential binding interface, which may be important for the microfibril assembly process and/or interactions with other microfibril components.
A correlation between the in vitro structural effects of amino acid substitutions with their cellular behaviour and consequences for intracellular trafficking and secretion is important for understanding the pathogenesis of MFS. Fibrillin‐1 biosynthesis, processing, and matrix deposition have been studied by pulse‐chase analyses of patient fibroblast cell cultures.44,45,218 The interpretation of such pulse‐chase studies, however, is complicated by the presence of normal fibrillin‐1 produced from the wild type allele, which cannot be distinguished from the mutant product. In order to study the fate of mutant fibrillin‐1, a recombinant system has been developed using a fibroblast host cell.219
In this system, fibrillin‐1 fragments containing two cysteine substitutions associated with classic MFS, C1117Y and C1129Y in cbEGF13, were retained intracellularly in the endoplasmic reticulum when expressed as a shortened form (100 kDa) of fibrillin‐1. This suggests that the delay in secretion observed in the patient cells is due to selective retention of mutant protein in the cell. In contrast, the G1127S folding substitution in the same domain was secreted into conditioned medium. This, together with the pulse‐chase studies of patient fibroblasts containing G1127S, which showed normal synthesis and secretion of fibrillin‐1 but reduced deposition in the extracellular matrix, suggests that this substitution has an extracellular dominant negative effect during or after incorporation of fibrillin‐1 into the microfibril. A greater disruption to cbEGF13 presumably results from the presence of an unpaired cysteine than from the localised structural effects of G1127S. Mutant proteins retained as a consequence of misfolding may result in functional haploinsufficiency or, alternatively, have an intracellular dominant negative effect. These functional studies of the structural changes introduced by missense mutations provide further insights into the pathogenic mechanisms leading to MFS.
FBN1 mutations and proteolysis
Most FBN1 missense mutations reported to date affect one of the highly conserved cysteine residues or a residue of the calcium‐binding consensus sequence in one of the 43 cbEGF‐like modules.149 Such mutations are predicted to reduce the calcium affinity of affected modules, which has been experimentally verified in a number of cases.212,213,220,221 On the other hand, calcium‐binding is important for the stability of cbEGF modules in a variety of ECM proteins such as fibrillin‐1,209,222,223,224 and removal of calcium by incubation leads to an increase of susceptibility to proteolysis.209,225,226,227 These findings have motivated several groups to investigate the effects of FBN1 mutations on the susceptibility of recombinant fibrillin fragments.
A number of FBN1 mutations have been shown to increase the susceptibility of recombinant fibrillin‐1 polypeptides to in vitro proteolysis (table 5). Many of the FBN1 mutations investigated in in vitro assays to date are predicted to reduce the calcium binding affinity of the cbEGF module harbouring the mutation. It has been shown by methods including two‐dimensional NMR spectroscopy216 and N‐terminal sequencing of mutant polypeptide proteolytic fragments,217 that FBN1 mutations can cause short and long range conformational deformations. The fact that FBN1 mutations can produce protease‐sensitive sites can be explained by reduced steric hindrance for the proteases caused by a missing calcium ion or other conformational changes.217 The molecular effects of calcium‐binding mutations may be influenced by the calcium‐binding affinity of affected cbEGF modules and by the location of the cbEGF module in the protein as well as the neighbouring modules.210 This, as well as the observation that different mutations within one cbEGF module can have a differential effect on susceptibility to proteolysis,228 provides potential explanations for some genotype‐phenotype correlations in MFS. Two mutations affecting the 1–3 disulfide bond of cbEGF30 of fibrillin‐1 had structural effects localised to the N‐terminal end of the mutant domain, with impaired domain packing with the preceding module.212 On the other hand, C750G, which affects the 5–6 disulfide bond of cbEGF7, increases proteolytic susceptibility of cbEGF8.214
Table 5 FBN1 mutations identified in individuals with MFS that have been shown to increase susceptibility to in vitro proteolysis.
| Mutation | Location | Reference |
|---|---|---|
| N548I | cbEGF4 | Reinhardt et al217 |
| R627C | cbEGF6 | Vollbrandt et al214 |
| C750G | cbEGF7 | Vollbrandt et al214 |
| E1073K | cbEGF12 | Reinhardt et al217 |
| G1127S | cbEGF13 | Whiteman et al216 |
| K1300E | cbEGF17 | Booms et al228 |
| C1320S | cbEGF17 | Booms et al228 |
| D1406G | cbEGF20 | Robinson and Booms229 |
| C1408F | cbEGF20 | Robinson and Booms229 |
| C1977R | cbEGF30 | Suk et al212 |
| C1977Y | cbEGF30 | Suk et al212 |
| N2183S | cbEGF33 | McGettrick et al210 |
The predicted amino acid change and the affected cbEGF modules are shown. Note that N2183S is a protein‐engineered mutation and has not been found in patients.
These data can be interpreted to mean that both the residues affected, the localisation within the cbEGF module, the localisation within the polypeptide chain of fibrillin‐1, and the nature of the neighbouring residues and the strength or flexibility of interdomain linkages all can be important for determining the effects of FBN1 mutations on proteolytic susceptibility.
While the above observations suggest that proteolysis may play a role in the pathogenesis of MFS, it should be noted that there is no formal proof of this available at the moment. Assuming that increased susceptibility to proteases does play a role in the pathogenesis of MFS, then there are at least two mechanisms that could be important. Proteolytic degradation of mutant fibrillin‐1 monomers could take place following secretion into the extracellular space but before incorporation into the microfibrils, thus reducing the total amount of fibrillin‐1 available for assembly of microfibrils. Alternatively, mutant fibrillin‐1 monomers could be incorporated into microfibrils and then represent a sort of Achilles' heel causing increased susceptibility of the entire microfibrillar structure to proteolysis and fragmentation.229 These issues, as well as a formal demonstration of the presence of fibrillin‐1 proteolytic fragments in tissues of MFS patients, represent potentially fruitful areas for future research.
Upregulation of matrix metalloproteinase expression by fibrillin fragments
The above observations suggest that FBN1 mutations might increase the susceptibility of entire microfibrils to proteolytic degradation, which could in turn lead to fragmentation of microfibrils in affected tissues. This hypothesis appears plausible because there is evidence of microfibrillar fragmentation in tissues of MFS patients230 and increased matrix metalloproteinase (MMP) concentrations in tissues of MFS patients,231,232 and fibrillin‐1 is susceptible to digestion by several MMPs.233 On the other hand, ECM proteins can influence cellular function and gene regulation by outside‐to‐inside signalling,234 and in some cases, fragments of ECM proteins acquire novel signalling properties that the intact molecules do not possess.235 For instance, increased concentrations of fibronectin fragments are found in osteoarthritis and can themselves upregulate the expression of several MMPs.236,237
A recent study showed that a recombinant fibrillin‐1 polypeptide containing the integrin binding RGD site of fibrillin‐1 can upregulate the expression and production of MMP‐1 and MMP‐3 in a cell culture system.238 A different recombinant fibrillin‐1 fragment containing a putative GxxPG elastin‐binding protein (EBP) interaction motif upregulates the expression and production of MMP‐1 by a factor of up to 9, an effect that was abolished by a mutation of the GxxPG consensus sequence.239 These observations provide a plausible explanation for the increased MMP concentrations seen in the tissues of MFS patients and suggest also the possibility of a vicious cycle whereby the constant presence of fibrillin‐1 fragments could lead to increased MMP production which in turn could cause the production of more fibrillin‐1 fragments.228 Additionally, since TGFβ1 can increase expression of several MMPs,240 increased protease‐mediated TGFβ release could further add to the proposed vicious cycle.
It has recently been shown that aortic extracts of the fibrillin‐1 underexpressing mgR/mgR MFS mouse model, as well as a recombinant fibrillin‐1 fragment containing a GxxPG EBP interaction motif, can stimulate macrophage chemotaxis, an effect that was significantly diminished by pretreatment of macrophages with lactose or with the elastin‐derived peptide VGVAPG, and by a mutation of the EBP recognition sequence in the fibrillin‐1 fragment, indicating involvement of EBP in mediating the effects (Gao and Robinson, under review). Additionally, investigation of macrophages in aortic specimens of Marfan patients demonstrated macrophage infiltration in the tunica media. These findings provide a plausible molecular mechanism for the inflammatory infiltrates observed in the mgR mouse model and suggest that inflammation may represent a component of the complex pathogenesis of MFS. Further work will be necessary to determine if inflammation and alterations of MMP regulation contribute in a significant way to the initiation or progression of disease.
Genetics of MFS in mouse models
Creation of genetically engineered mouse models of MFS has been instrumental in modifying our long‐held belief that MFS is a classic dominant‐negative disorder, in which the abnormal protein derived from the mutant allele functionally neutralises the normal protein produced from the wild type allele. These animal studies have revealed unsuspected functions of extracellular microfibrils in orchestrating cellular activities and morphogenetic programs through the modulation of critical signalling events. Most importantly, they have opened exciting new opportunities for the development of productive treatment strategies in MFS, in addition to implicating dysregulated TGFβ signalling in the genesis of other developmentally and postnatally acquired elastin‐deficient states.
The first new insight regarding the pathogenesis of cardiovascular disease derived from the study of mouse strains homozygous for hypomorphic (low expressing) fibrillin‐1 (Fbn1) alleles. They included the mgΔ/mgΔ mice, which express a centrally deleted protein at about 5% of the normal level and die shortly after birth, and mgR/mgR mice, which express full length protein at about 15% of the normal level and die at 3–6 months of age.10,11,241 That mgΔ/mgΔ mice exhibit only focal disruptions in elastic fibre organisation demonstrated that minimal (if any) residual microfibrillar function is sufficient to support the deposition of extended elastic structures.10 On the other hand, the longer life span and normal aortic wall architecture of mgR/mgR mice allowed demonstration of the pathogenic sequence that culminates in aneurysm in these animals.11 The secondary events include regional elastic fibre calcification, intimal hyperplasia, and adventitial inflammation, findings that were generally spatially coincident and which were subsequently confirmed in MFS patients as well.241 Loss of physical connections between vascular smooth muscle cells and neighbouring elastic fibres that are normally mediated by fibrillin‐1 are believed to contribute to the initiation of this destructive process.241 As a result, the morphology and synthetic program of the cells are altered, perhaps in an abortive attempt to reconstitute a normal extracellular environment, and the ensuing remodelling process is characterised by overproduction of multiple structural matrix components and mediators of elastolysis, including MMPs 2 and 9.241 Subsequent breach of the internal or external elastic laminae allows infiltration of inflammatory cells into the media, resulting in intense elastolysis that correlates temporally with aneurysm formation and dissection. The critical insights derived from this work are that a normal complement of fibrillin‐1 is needed for elastic fibre homeostasis rather than formation, and that there may be important opportunities to alter the natural history of disease through modification of secondary events including inflammation and matrix destruction.
Haploinsufficiency contributes to disease progression
The above data suggested a threshold hypothesis, whereby the relative abundance of functionally competent fibrillin‐1 microfibrils determines the clinical severity of the MFS phenotype. Consistent with this postulate, recent evidence indicated that half‐normal production of wild type fibrillin‐1 (haploinsufficiency) can contribute to reaching the threshold loss of microfibril function needed for clinical expression of MFS.242 First, transgenic overexpression of mutant protein, in the context of two normal Fbn1 alleles, was insufficient to cause the vascular changes of MFS seen in mice heterozygous for a comparable missense mutation (C1039G). Second, mice heterozygous for a null Fbn1 allele showed similar alterations in aortic wall architecture as Fbn1C1039G/+ mice. Finally, the transgenic addition of an additional wild type allele to C1039G mice rescued the aortic phenotype. Thus, a normal complement of fibrillin‐1 may be needed to initiate productive microfibrillar assembly.242 While the haploinsufficiency‐imposed reduction in microfibril abundance may or may not be sufficient, in isolation, to lead to classic MFS, it appears critical to the context within which a dominant negative effect can achieve clinical significance. These data raised the possibility that boosting fibrillin‐1 expression may be a productive therapeutic strategy and thus offered attractive hypotheses regarding the role of genetic modifiers in the modulation of disease severity.
Microfibrillar modulation of signalling molecules
More recent observations have indicated that microfibril‐associated disorders are actually developmental abnormalities that affect multiple morphogenetic programs and become clinically manifest later in life.12,243,244 Multifunctional molecules that are secreted into the extracellular space and which signal through membrane‐bound receptors are the primary triggers and modulators of cellular activities underlying development and growth, and tissue homeostasis and repair.245 Findings in fibrillin deficient mice have implicated the participation of extracellular microfibrils in TGF/BMP signalling events that control patterning and morphogenesis. In turn, these results have suggested functional coupling between tissue‐specific organisation of elastic fibre macroaggregates, and their ability to perform instructive as well as structural functions.
The first evidence implicating fibrillin‐rich microfibrils in developmental process derived from the finding that fibrillin‐2 (Fbn2) mice display bilateral syndactyly, as a result of failed BMP‐induced apoptosis of the prospective interdigital mesenchyme.12 The patterning defect was associated with altered microfibril assembly only and specifically in the developing autopod, and with loss of interdigital tissue competence to respond to apoptotic cues from exogenously administered BMP signals. Importantly, the combination of Fbn2 and Bmp7 haploinsufficiency, which are by themselves phenotypically silent, resulted in impaired digit formation in the absence of additional manifestation. These data therefore suggested that the tissue‐specific architecture of fibrillin‐rich microfibrils may positively regulate BMP signalling by concentrating effector molecules at the site of intended function.
The other evidence for a direct role of the fibrillins in the regulation of signalling molecules derived from the study of lung disease in mgΔ and mgR mice.243 A distinct subset of individuals with MFS have clinically manifest lung disease, generally presenting as spontaneous pneumothorax, and even more of them show radiographic evidence of obstructive lung disease, including upper lobe bullae and/or more diffuse airspace widening.246,247 Such findings have been traditionally reconciled with models that invoke structural failure of the connective tissue with subsequent inflammation and destructive emphysematous changes. However, mgΔ/mgΔ mice displayed homogeneous widening of the distal alveolar saccules without any evidence of inflammation or tissue destruction.243 Instead, a dramatic paucity of primordial alveolar septa was the predominant finding, suggesting a primary developmental failure of distal alveolar septation rather than classic emphysema, as previously inferred. Failure of lung septation in the mutant mice was correlated temporally and spatially with excess immunoreactive free TGFβ resulting from increased local activation rather than production and secretion of the cytokine.243 More importantly, TGFβ neutralising antibodies rescued lung morphogenesis in mgΔ/mgΔ mice. These results raised the possibility that a similar mechanism may underlie other manifestations of MFS, including bone overgrowth, valve changes, and aortic dilatation.243 Indeed, myxomatous changes of the mitral valve in Fbn1C1039G/+ mice were recently correlated with excess TGFβ signalling and were prevented by TGFβ antagonism in vivo.244
Dysregulated TGFβ signalling in MFS‐like conditions
A recent report suggested heterozygous loss of function mutations in the type II TGFβ receptor (TβRII) phenocopy MFS.111 Selected mutant receptors were shown to be incapable of propagating TGFβ signal when expressed in cells naive for TβRII. A subsequent report suggested that the phenotype associated with heterozygous mutations in either TβRI or TβRII is distinct from MFS, but shows some overlap including aortic aneurysm, arachnodactyly, pectus deformity, and scoliosis.112 Highly prevalent distinguishing features included cleft palate/bifid uvula, hypertelorism, and arterial tortuosity; more variable features included craniosynostosis, neurocognitive abnormalities, and congenital heart disease. Importantly, heterozygous patient cells showed full preservation of the acute phase response to TGFβ while patient‐derived tissues showed evidence of increased (rather than decreased) TGFβ signalling.112 While adding to the complexity, these data support the contentions that many features of microfibril disorders likely manifest failure of proper regulation of cytokine function, and that consideration of both primary and secondary events will be required to attain full mechanistic insight. Efforts are underway to comprehensively evaluate the utility of TGFβ antagonism in the treatment of the multisystem manifestations of MFS, an initiative that will require both animal studies and multicentre collaborative human clinical trials. There is particular attractiveness for consideration of the use of angiotensin II type I receptor blockers, such as losartan, that both lower blood pressure (a known positive effect in MFS) and lead to a clinically relevant attenuation of TGFβ signalling.
Conclusions
The past 5 years have witnessed remarkable progress in elucidating the anatomy and physiology of the microfibrils and in understanding the pathogenesis of MFS on a molecular level. In particular, our understanding of the pathophysiology of MFS has evolved away from previous more or less static concepts involving a dominant negative mechanism resulting in structural weakness of connective tissue.
A full understanding of the pathogenesis of MFS will require further advances in many different areas. We are beginning to understand a range of pathophysiological processes involving protein folding and stability, microfibrillar assembly, proteolysis of mutant fibrillin monomers, secondary effects of fragmentation, loss of smooth muscle cell attachments, and alterations in TGFβ signalling. The challenge for the coming years will be to sort out the relative contributions of these defects to the pathogenesis of the various manifestations of MFS. There is no reason to believe that the molecular pathogenesis of ectopia lentis must be the same as that of long bone overgrowth or aortic dissection, and indeed the fact that the various manifestations of MFS do not always occur together in the same patient suggests that there may be important differences in detailed pathogenetic mechanisms.
We still do not know enough about the molecular mechanisms in MFS to develop a model that would explain the sequence of events leading from a mutation in FBN1 to clinical disease. Clinical observations and experimental studies using in vitro systems, cell culture models, or animal models have demonstrated multiple abnormalities, but it is not entirely clear how the sequence of pathogenetic events seen in MFS occurs. One interpretation of the available evidence is that FBN1 mutations lead to either a microfibrillar assembly defect with reduced levels of fibrillin‐rich microfibrils in tissues, or that mutant fibrillin monomers are incorporated into microfibrils and cause a slow but progressive loss of microfibrils. By whatever mechanism this occurs, the loss of fibrillin‐rich microfibrils then has a series of other secondary effects, including a disturbance of the ability to sequester TGFβ and modulate its activity appropriately. Disturbances in the ability of microfibrils to adequately sequester other growth factors may also be important. Increased levels of MMPs may be a result of increased TGFβ activity, of phenotypic alteration of vascular smooth muscle cells owing to loss of cell attachments that are normally mediated by fibrillin‐1, or of effects of the fibrillin fragments themselves. Other secondary effects of fibrillin or elastin fragments could include recruitment of macrophages into the aortic wall by chemotactic stimulation, which could contribute to further progression of aneurysm formation.
The study of mutations and genotype‐phenotype correlations in MFS and related disorders may provide clues that will help elucidate the above issues. At present, there are no plausible molecular explanations for most of the genotype‐phenotype correlations that have been observed with FBN1 mutations. For instance, we are not able to explain why a mutation in exon 59 is associated with isolated ectopia lentis132 and why other mutations are associated with the severe neonatal form of MFS.129
Although our knowledge of the molecular pathogenesis of MFS is still incomplete, the new paradigms emerging from recent studies have the potential to improve treatment for individuals with MFS. The most promising new direction is the pharmacological manipulation of the TGFβ signalling pathway with angiotensin II type 1 receptor blockers such as losartan, which both lowers blood pressure and attenuates TGFβ signalling.248
Although judgments concerning the usefulness and indications of losartan will have to await the results of clinical trials, it does appear certain that our current understanding of MFS as a no‐longer purely structural disorder of the matrix will reveal new molecular targets that can be exploited pharmacologically to improve the lives of individuals with this disorder.
Acknowledgements
The authors would like to thank the anonymous reviewers for very useful suggestions.
Electronic‐database information
The UMD FBN1 database is at http://www.umd.be.
Abbreviations
BMP - bone morphogenetic protein
cbEGF - calcium binding epidermal growth factor
CCA - congenital contractural arachnodactyly
CMN - cystic medial necrosis
CSGE - conformation sensitive gel electrophoresis
DHPLC - denaturing high performance liquid chromatography
EBP - elastin‐binding protein
ECM - extracellular matrix
HNPCC - hereditary non‐polyposis colorectal cancer
LAP - latency‐associated peptide
LDS - Loeys‐Dietz aortic aneurysm syndrome
LLC - large latent complexes
LTBP - latent‐TGFβ‐binding protein
MAGP‐1 - microfibril‐associated glycoprotein‐1
MFS - Marfan syndrome
MFS2 - type 2 Marfan syndrome
MMP - matrix metalloproteinase
MMR - mismatch repair
MSI - microsatellite instability
NMR - nuclear magnetic resonance
PTC - premature termination codon
SSCP - single stranded conformation polymorphism
TAAD - thoracic ascending aortic aneurysms and dissections
TβRII - type II TGFβ receptor
TGFβ - transforming growth factor‐β
UMD - Universal Mutation Database
WMS - Weill‐Marchesani syndrome
Footnotes
The authors of this work would like to acknowledge support from Marfan Hilfe (Deutschland) e.V., the Canadian Marfan Association, the National Marfan Foundation (USA), the Deutsche Forschungsgemeinschaft (Grant SFB367‐A1), the Canadian Institutes of Health Research (Grant MOP‐68836), the Fund for Scientific Research Flanders, the Howard Hughes Medical Institute, National Institutes of Health (AR‐41135, AR‐049698 and AR‐42044), the St Giles Foundation, the James D. Farley Family, the Smilow Center for Marfan Syndrome Research, and the Medical Research Council (G000164)
Competing interests: none declared
The UMD FBN1 database is at http://www.umd.be.
References
- 1.Robinson P N, Godfrey M.Marfan syndrome: a primer for clinicians and scientists. New York: Kluwer Academic/Plenum, 2004
- 2.Pyeritz R E. The Marfan syndrome. Annu Rev Med 200051481–510. [DOI] [PubMed] [Google Scholar]
- 3.Robinson P N, Godfrey M. The molecular genetics of Marfan syndrome and related microfibrillopathies. J Med Genet 200037(1)9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Apfelroth S D, Hu W, Davis E C, Sanguineti C, Bonadio J, Mecham R P, Ramirez F. Structure and expression of fibrillin‐2, a novel microfibrillar component preferentially located in elastic matrices. J Cell Biol 1994124(5)855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corson G M, Charbonneau N L, Keene D R, Sakai L Y. Differential expression of fibrillin‐3 adds to microfibril variety in human and avian, but not rodent, connective tissues. Genomics 200483(3)461–472. [DOI] [PubMed] [Google Scholar]
- 6.Carta L, Pereira L, Arteaga‐Solis E, Lee‐Arteaga S Y, Lenart B, Starcher B, Merkel C A, Sukoyan M, Kerkis A, Hazeki N, Keene D R, Sakai L Y, Ramirez F. Fibrillins 1 and 2 perform partially overlapping functions during aortic development. J Biol Chem 20062818016–8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakai L Y, Keene D R, Glanville R W, Bachinger H P. Purification and partial characterization of fibrillin, a cysteine‐rich structural component of connective tissue microfibrils. J Biol Chem 1991266(22)14763–14770. [PubMed] [Google Scholar]
- 8.Charbonneau N L, Dzamba B J, Ono R N, Keene D R, Corson G M, Reinhardt D P, Sakai L Y. Fibrillins can co‐assemble in fibrils, but fibrillin fibril composition displays cell‐specific differences. J Biol Chem 2003278(4)2740–2749. [DOI] [PubMed] [Google Scholar]
- 9.Gallagher B C, Sakai L Y, Little C D. Fibrillin delineates the primary axis of the early avian embryo. Dev Dyn 1993196(1)70–78. [DOI] [PubMed] [Google Scholar]
- 10.Pereira L, Andrikopoulos K, Tian J, Lee S Y, Keene D R, Ono R, Reinhardt D P, Sakai L Y, Biery N J, Bunton T, Dietz H C, Ramirez F. Targetting of the gene encoding fibrillin‐1 recapitulates the vascular aspect of Marfan syndrome. Nat Genet 199717(2)218–222. [DOI] [PubMed] [Google Scholar]
- 11.Pereira L, Lee S Y, Gayraud B, Andrikopoulos K, Shapiro S D, Bunton T, Biery N J, Dietz H C, Sakai L Y, Ramirez F. Pathogenetic sequence for aneurysm revealed in mice underexpressing fibrillin‐1. Proc Natl Acad Sci U S A 199996(7)3819–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arteaga‐Solis E, Gayraud B, Lee S Y, Shum L, Sakai L, Ramirez F. Regulation of limb patterning by extracellular microfibrils. J Cell Biol 2001154(2)275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oklu R, Hesketh R. The latent transforming growth factor beta binding protein (LTBP) family. Biochem J 2000352(Pt 3)601–610. [PMC free article] [PubMed] [Google Scholar]
- 14.Rifkin D B. Latent transforming growth factor‐beta (TGF‐beta) binding proteins: orchestrators of TGF‐beta availability. J Biol Chem 2005280(9)7409–7412. [DOI] [PubMed] [Google Scholar]
- 15.Todorovic V, Jurukovski V, Chen Y, Fontana L, Dabovic B, Rifkin D B. Latent TGF‐beta binding proteins. Int J Biochem Cell Biol 200537(1)38–41. [DOI] [PubMed] [Google Scholar]
- 16.Handford P A, Mayhew M, Baron M, Winship P R, Campbell I D, Brownlee G G. Key residues involved in calcium‐binding motifs in EGF‐like domains. Nature 1991351(6322)164–167. [DOI] [PubMed] [Google Scholar]
- 17.Yuan X, Downing A K, Knott V, Handford P A. Solution structure of the transforming growth factor beta‐binding protein‐like module, a domain associated with matrix fibrils. EMBO J 199716(22)6659–6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gleizes P E, Beavis R C, Mazzieri R, Shen B, Rifkin D B. Identification and characterization of an eight‐cysteine repeat of the latent transforming growth factor‐beta binding protein‐1 that mediates bonding to the latent transforming growth factor‐beta1. J Biol Chem 1996271(47)29891–29896. [DOI] [PubMed] [Google Scholar]
- 19.Saharinen J, Taipale J, Keski‐Oja J. Association of the small latent transforming growth factor‐beta with an eight cysteine repeat of its binding protein LTBP‐1. EMBO J 199615(2)245–253. [PMC free article] [PubMed] [Google Scholar]
- 20.Saharinen J, Keski‐Oja J. Specific sequence motif of 8‐Cys repeats of TGF‐beta binding proteins, LTBPs, creates a hydrophobic interaction surface for binding of small latent TGF‐beta. Mol Biol Cell 200011(8)2691–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereira L, D'Alessio M, Ramirez F, Lynch J R, Sykes B, Pangilinan T, Bonadio J. Genomic organization of the sequence coding for fibrillin, the defective gene product in Marfan syndrome. Hum Mol Genet 19932(10)1762. [DOI] [PubMed] [Google Scholar]
- 22.Reinhardt D P, Gambee J E, Ono R N, Bachinger H P, Sakai L Y. Initial steps in assembly of microfibrils. Formation of disulfide‐cross‐linked multimers containing fibrillin‐1. J Biol Chem 2000275(3)2205–2210. [DOI] [PubMed] [Google Scholar]
- 23.Giltay R, Timpl R, Kostka G. Sequence, recombinant expression and tissue localization of two novel extracellular matrix proteins, fibulin‐3 and fibulin‐4. Matrix Biol 199918(5)469–480. [DOI] [PubMed] [Google Scholar]
- 24.Marson A, Rock M J, Cain S A, Freeman L J, Morgan A, Mellody K, Shuttleworth C A, Baldock C, Kielty C M. Homotypic fibrillin‐1 interactions in microfibril assembly. J Biol Chem 2005280(6)5013–5021. [DOI] [PubMed] [Google Scholar]
- 25.Kielty C M, Wess T J, Haston J L, Sherratt M J, Baldock C, Shuttleworth C A, Robinson P N, Godfrey M. Organisation and biomechanical properties of fibrillin microfibrils. In: eds. Marfan syndrome: a primer for clinicians and scientists. New York: Kluwer Academic/Plenum, 2004130–142.
- 26.Kielty C M, Sherratt M J, Shuttleworth C A. Elastic fibres. J Cell Sci 2002115(Pt 14)2817–2828. [DOI] [PubMed] [Google Scholar]
- 27.Keene D R, Maddox B K, Kuo H J, Sakai L Y, Glanville R W. Extraction of extendable beaded structures and their identification as fibrillin‐containing extracellular matrix microfibrils. J Histochem Cytochem 199139(4)441–449. [DOI] [PubMed] [Google Scholar]
- 28.Baldock C, Koster A J, Ziese U, Rock M J, Sherratt M J, Kadler K E, Shuttleworth C A, Kielty C M. The supramolecular organization of fibrillin‐rich microfibrils. J Cell Biol 2001152(5)1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis E C, Roth R A, Heuser J E, Mecham R P. Ultrastructural properties of ciliary zonule microfibrils. J Struct Biol 2002139(2)65–75. [DOI] [PubMed] [Google Scholar]
- 30.Qian R Q, Glanville R W. Alignment of fibrillin molecules in elastic microfibrils is defined by transglutaminase‐derived cross‐links. Biochemistry 199736(50)15841–15847. [DOI] [PubMed] [Google Scholar]
- 31.Cain S A, Morgan A, Sherratt M J, Ball S G, Shuttleworth C A, Kielty C M. Proteomic analysis of fibrillin‐rich microfibrils. Proteomics 20066(1)111–122. [DOI] [PubMed] [Google Scholar]
- 32.Baldock C, Gilpin C J, Koster A J, Ziese U, Kadler K E, Kielty C M, Holmes D F. Three‐dimensional reconstructions of extracellular matrix polymers using automated electron tomography. J Struct Biol 2002138(1–2)130–136. [DOI] [PubMed] [Google Scholar]
- 33.Sherratt M J, Baldock C, Haston J L, Holmes D F, Jones C J, Shuttleworth C A, Wess T J, Kielty C M. Fibrillin microfibrils are stiff reinforcing fibres in compliant tissues. J Mol Biol 2003332(1)183–193. [DOI] [PubMed] [Google Scholar]
- 34.Kielty C M, Sherratt M J, Marson A, Baldock C, Parry D, Squire J. Fibrillin microfibrils. In: eds. Advances in protein chemistry. Vol 70. San Diego: Academic Press, 2005405–436. [DOI] [PubMed]
- 35.Downing A K, Knott V, Werner J M, Cardy C M, Campbell I D, Handford P A. Solution structure of a pair of calcium‐binding epidermal growth factor‐like domains: implications for the Marfan syndrome and other genetic disorders. Cell 199685(4)597–605. [DOI] [PubMed] [Google Scholar]
- 36.Lee S S, Knott V, Jovanovic J, Harlos K, Grimes J M, Choulier L, Mardon H J, Stuart D I, Handford P A. Structure of the integrin binding fragment from fibrillin‐1 gives new insights into microfibril organization. Structure (Camb) 200412(4)717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen S A, Corbett A R, Knott V, Redfield C, Handford P A. Ca2+‐dependent interface formation in fibrillin‐1. J Biol Chem 2005280(14)14076–14084. [DOI] [PubMed] [Google Scholar]
- 38.Kielty C M, Wess T J, Haston L, Ashworth J L, Sherratt M J, Shuttleworth C A. Fibrillin‐rich microfibrils: elastic biopolymers of the extracellular matrix. J Muscle Res Cell Motil 200223(5–6)581–596. [PubMed] [Google Scholar]
- 39.Godfrey M, Olson S, Burgio R G, Martini A, Valli M, Cetta G, Hori H, Hollister D W. Unilateral microfibrillar abnormalities in a case of asymmetric Marfan syndrome. Am J Hum Genet 199046(4)661–671. [PMC free article] [PubMed] [Google Scholar]
- 40.Godfrey M, Menashe V, Weleber R G, Koler R D, Bigley R H, Lovrien E, Zonana J, Hollister D W. Cosegregation of elastin‐associated microfibrillar abnormalities with the Marfan phenotype in families. Am J Hum Genet 199046(4)652–660. [PMC free article] [PubMed] [Google Scholar]
- 41.Hollister D W, Godfrey M, Sakai L Y, Pyeritz R E. Immunohistologic abnormalities of the microfibrillar‐fiber system in the Marfan syndrome. N Engl J Med 1990323(3)152–159. [DOI] [PubMed] [Google Scholar]
- 42.Halliday D, Hutchinson S, Kettle S, Firth H, Wordsworth P, Handford P A. Molecular analysis of eight mutations in FBN1. Hum Genet 1999105(6)587–597. [DOI] [PubMed] [Google Scholar]
- 43.Milewicz D M, Pyeritz R E, Crawford E S, Byers P H. Marfan syndrome: defective synthesis, secretion, and extracellular matrix formation of fibrillin by cultured dermal fibroblasts. J Clin Invest 199289(1)79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aoyama T, Tynan K, Dietz H C, Francke U, Furthmayr H. Missense mutations impair intracellular processing of fibrillin and microfibril assembly in Marfan syndrome. Hum Mol Genet 19932(12)2135–2140. [DOI] [PubMed] [Google Scholar]
- 45.Aoyama T, Francke U, Dietz H C, Furthmayr H. Quantitative differences in biosynthesis and extracellular deposition of fibrillin in cultured fibroblasts distinguish five groups of Marfan syndrome patients and suggest distinct pathogenetic mechanisms. J Clin Invest 199494(1)130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brenn T, Aoyama T, Francke U, Furthmayr H. Dermal fibroblast culture as a model system for studies of fibrillin assembly and pathogenetic mechanisms: defects in distinct groups of individuals with Marfan's syndrome. Lab Invest 199675(3)389–402. [PubMed] [Google Scholar]
- 47.Kielty C M, Shuttleworth C A. Abnormal fibrillin assembly by dermal fibroblasts from two patients with Marfan syndrome. J Cell Biol 1994124(6)997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kielty C M, Phillips J E, Child A H, Pope F M, Shuttleworth C A. Fibrillin secretion and microfibril assembly by Marfan dermal fibroblasts. Matrix Biol 199414(2)191–199. [DOI] [PubMed] [Google Scholar]
- 49.Kielty C M, Davies S J, Phillips J E, Jones C J, Shuttleworth C A, Charles S J. Marfan syndrome: fibrillin expression and microfibrillar abnormalities in a family with predominant ocular defects. J Med Genet 199532(1)1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Milewicz D M, Grossfield J, Cao S N, Kielty C, Covitz W, Jewett T. A mutation in FBN1 disrupts profibrillin processing and results in isolated skeletal features of the Marfan syndrome. J Clin Invest 199595(5)2373–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reinhardt D P, Keene D R, Corson G M, Poschl E, Bachinger H P, Gambee J E, Sakai L Y. Fibrillin‐1: organization in microfibrils and structural properties. J Mol Biol 1996258(1)104–116. [DOI] [PubMed] [Google Scholar]
- 52.Lonnqvist L, Reinhardt D, Sakai L, Peltonen L. Evidence for furin‐type activity‐mediated C‐terminal processing of profibrillin‐1 and interference in the processing by certain mutations. Hum Mol Genet 19987(13)2039–2044. [DOI] [PubMed] [Google Scholar]
- 53.Raghunath M, Putnam E A, Ritty T, Hamstra D, Park E S, Tschodrich‐Rotter M, Peters R, Rehemtulla A, Milewicz D M. Carboxy‐terminal conversion of profibrillin to fibrillin at a basic site by PACE/furin‐like activity required for incorporation in the matrix. J Cell Sci 1999112(Pt 7)1093–1100. [DOI] [PubMed] [Google Scholar]
- 54.Ritty T M, Broekelmann T, Tisdale C, Milewicz D M, Mecham R P. Processing of the fibrillin‐1 carboxyl‐terminal domain. J Biol Chem 1999274(13)8933–8940. [DOI] [PubMed] [Google Scholar]
- 55.Wright D W, Mayne R. Vitreous humor of chicken contains two fibrillar systems: an analysis of their structure. J Ultrastruct Mol Struct Res 1988100(3)224–234. [DOI] [PubMed] [Google Scholar]
- 56.Wallace R N, Streeten B W, Hanna R B. Rotary shadowing of elastic system microfibrils in the ocular zonule, vitreous, and ligamentum nuchae. Curr Eye Res 199110(1)99–109. [DOI] [PubMed] [Google Scholar]
- 57.Lin G, Tiedemann K, Vollbrandt T, Peters H, Batge B, Brinckmann J, Reinhardt D P. Homo‐ and heterotypic fibrillin‐1 and ‐2 interactions constitute the basis for the assembly of microfibrils. J Biol Chem 2002277(52)50795–50804. [DOI] [PubMed] [Google Scholar]
- 58.Trask T M, Ritty T M, Broekelmann T, Tisdale C, Mecham R P. N‐terminal domains of fibrillin 1 and fibrillin 2 direct the formation of homodimers: a possible first step in microfibril assembly. Biochem J 1999340(Pt 3)693–701. [PMC free article] [PubMed] [Google Scholar]
- 59.Ashworth J L, Kelly V, Wilson R, Shuttleworth C A, Kielty C M. Fibrillin assembly: dimer formation mediated by amino‐terminal sequences. J Cell Sci 1999112(Pt 20)3549–3558. [DOI] [PubMed] [Google Scholar]
- 60.Kielty C M, Shuttleworth C A. Synthesis and assembly of fibrillin by fibroblasts and smooth muscle cells. J Cell Sci 1993106(Pt 1)167–173. [DOI] [PubMed] [Google Scholar]
- 61.Bowness J M, Tarr A H. epsilon(gamma‐Glutamyl)lysine crosslinks are concentrated in a non‐collagenous microfibrillar fraction of cartilage. Biochem Cell Biol 199775(1)89–91. [DOI] [PubMed] [Google Scholar]
- 62.Thurmond F A, Koob T J, Bowness J M, Trotter J A. Partial biochemical and immunologic characterization of fibrillin microfibrils from sea cucumber dermis. Connect Tissue Res 199736(3)211–222. [DOI] [PubMed] [Google Scholar]
- 63.Brown‐Augsburger P, Broekelmann T, Mecham L, Mercer R, Gibson M A, Cleary E G, Abrams W R, Rosenbloom J, Mecham R P. Microfibril‐associated glycoprotein binds to the carboxyl‐terminal domain of tropoelastin and is a substrate for transglutaminase. J Biol Chem 1994269(45)28443–28449. [PubMed] [Google Scholar]
- 64.Raghunath M, Cankay R, Kubitscheck U, Fauteck J D, Mayne R, Aeschlimann D, Schlotzer‐Schrehardt U. Transglutaminase activity in the eye: cross‐linking in epithelia and connective tissue structures. Invest Ophthalmol Vis Sci 199940(12)2780–2787. [PubMed] [Google Scholar]
- 65.Thurmond F, Trotter J. Morphology and biomechanics of the microfibrillar network of sea cucumber dermis. J Exp Biol 1996199(Pt 8)1817–1828. [DOI] [PubMed] [Google Scholar]
- 66.Tiedemann K, Batge B, Muller P K, Reinhardt D P. Interactions of fibrillin‐1 with heparin/heparan sulfate, implications for microfibrillar assembly. J Biol Chem 2001276(38)36035–36042. [DOI] [PubMed] [Google Scholar]
- 67.Ritty T M, Broekelmann T J, Werneck C C, Mecham R P. Fibrillin‐1 and ‐2 contain heparin‐binding sites important for matrix deposition and that support cell attachment. Biochem J 2003375(Pt 2)425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cain S A, Baldock C, Gallagher J, Morgan A, Bax D V, Weiss A S, Shuttleworth C A, Kielty C M. Fibrillin‐1 interactions with heparin: implications for microfibril and elastic fiber assembly. J Biol Chem 2005280(34)30526–30537. [DOI] [PubMed] [Google Scholar]
- 69.Tiedemann K, Sasaki T, Gustafsson E, Gohring W, Batge B, Notbohm H, Timpl R, Wedel T, Schlotzer‐Schrehardt U, Reinhardt D P. Microfibrils at basement membrane zones interact with perlecan via fibrillin‐1. J Biol Chem 2005280(12)11404–11412. [DOI] [PubMed] [Google Scholar]
- 70.Gibson M A, Robinson P N, Godfrey M. Microfibril‐associated glycoprotein‐1 (MAGP‐1) and other non‐fibrillin macromolecules which may possess a functional association with the 10 nm microfibrils. In: eds. Marfan syndrome: a primer for clinicians and scientists. New York: Kluwer Academic/Plenum, 2005161–178.
- 71.Gibson M A, Hughes J L, Fanning J C, Cleary E G. The major antigen of elastin‐associated microfibrils is a 31‐kDa glycoprotein. J Biol Chem 1986261(24)11429–11436. [PubMed] [Google Scholar]
- 72.Henderson M, Polewski R, Fanning J C, Gibson M A. Microfibril‐associated glycoprotein‐1 (MAGP‐1) is specifically located on the beads of the beaded‐filament structure for fibrillin‐containing microfibrils as visualized by the rotary shadowing technique. J Histochem Cytochem 199644(12)1389–1397. [DOI] [PubMed] [Google Scholar]
- 73.Gibson M A, Finnis M L, Kumaratilake J S, Cleary E G. Microfibril‐associated glycoprotein‐2 (MAGP‐2) is specifically associated with fibrillin‐containing microfibrils but exhibits more restricted patterns of tissue localization and developmental expression than its structural relative MAGP‐1. J Histochem Cytochem 199846(8)871–886. [DOI] [PubMed] [Google Scholar]
- 74.Penner A S, Rock M J, Kielty C M, Shipley J M. Microfibril‐associated glycoprotein‐2 interacts with fibrillin‐1 and fibrillin‐2 suggesting a role for MAGP‐2 in elastic fiber assembly. J Biol Chem 2002277(38)35044–35049. [DOI] [PubMed] [Google Scholar]
- 75.Rock M J, Cain S A, Freeman L J, Morgan A, Mellody K, Marson A, Shuttleworth C A, Weiss A S, Kielty C M. Molecular basis of elastic fiber formation. Critical interactions and a tropoelastin‐fibrillin‐1 cross‐link. J Biol Chem 2004279(22)23748–23758. [DOI] [PubMed] [Google Scholar]
- 76.Jensen S A, Reinhardt D P, Gibson M A, Weiss A S. Protein interaction studies of MAGP‐1 with tropoelastin and fibrillin‐1. J Biol Chem 2001276(43)39661–39666. [DOI] [PubMed] [Google Scholar]
- 77.Reinboth B, Hanssen E, Cleary E G, Gibson M A. Molecular interactions of biglycan and decorin with elastic fiber components: biglycan forms a ternary complex with tropoelastin and microfibril‐associated glycoprotein 1. J Biol Chem 2002277(6)3950–3957. [DOI] [PubMed] [Google Scholar]
- 78.Tsuruga E, Yajima T, Irie K. Microfibril‐associated glycoprotein‐1 and fibrillin‐2 are associated with tropoelastin deposition in vitro. Int J Biochem Cell Biol 200537(1)120–129. [DOI] [PubMed] [Google Scholar]
- 79.Finnis M L, Gibson M A. Microfibril‐associated glycoprotein‐1 (MAGP‐1) binds to the pepsin‐resistant domain of the alpha3(VI) chain of type VI collagen. J Biol Chem 1997272(36)22817–22823. [DOI] [PubMed] [Google Scholar]
- 80.Nehring L C, Miyamoto A, Hein P W, Weinmaster G, Shipley J M. The extracellular matrix protein MAGP‐2 interacts with Jagged1 and induces its shedding from the cell surface. J Biol Chem 2005280(21)20349–20355. [DOI] [PubMed] [Google Scholar]
- 81.Charbonneau N L, Ono R N, Corson G M, Keene D R, Sakai L Y. Fine tuning of growth factor signals depends on fibrillin microfibril networks. Birth Defects Res C Embryo Today 200472(1)37–50. [DOI] [PubMed] [Google Scholar]
- 82.Gregory K E, Ono R N, Charbonneau N L, Kuo C L, Keene D R, Bachinger H P, Sakai L Y. The prodomain of BMP‐7 targets the BMP‐7 complex to the extracellular matrix. J Biol Chem 2005280(30)27970–27980. [DOI] [PubMed] [Google Scholar]
- 83.Homandberg G A, Hui F, Wen C, Purple C, Bewsey K, Koepp H, Huch K, Harris A. Fibronectin‐fragment‐induced cartilage chondrolysis is associated with release of catabolic cytokines. Biochem J 1997321(Pt 3)751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brassart B, Randoux A, Hornebeck W, Emonard H. Regulation of matrix metalloproteinase‐2 (gelatinase A, MMP‐2), membrane‐type matrix metalloproteinase‐1 (MT1‐MMP) and tissue inhibitor of metalloproteinases‐2 (TIMP‐2) expression by elastin‐derived peptides in human HT‐1080 fibrosarcoma cell line. Clin Exp Metastasis 199816(6)489–500. [DOI] [PubMed] [Google Scholar]
- 85.Brassart B, Fuchs P, Huet E, Alix A J, Wallach J, Tamburro A M, Delacoux F, Haye B, Emonard H, Hornebeck W, Debelle L. Conformational dependence of collagenase (matrix metalloproteinase‐1) up‐regulation by elastin peptides in cultured fibroblasts. J Biol Chem 2001276(7)5222–5227. [DOI] [PubMed] [Google Scholar]
- 86.Kagan H M, Vaccaro C A, Bronson R E, Tang S S, Brody J S. Ultrastructural immunolocalization of lysyl oxidase in vascular connective tissue. J Cell Biol 1986103(3)1121–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Somerville R P, Jungers K A, Apte S S. Discovery and characterization of a novel, widely expressed metalloprotease, ADAMTS10, and its proteolytic activation. J Biol Chem 2004279(49)51208–51217. [DOI] [PubMed] [Google Scholar]
- 88.Dagoneau N, Benoist‐Lasselin C, Huber C, Faivre L, Megarbane A, Alswaid A, Dollfus H, Alembik Y, Munnich A, Legeai‐Mallet L, Cormier‐Daire V. ADAMTS10 mutations in autosomal recessive Weill‐Marchesani syndrome. Am J Hum Genet 200475(5)801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clarke A W, Weiss A S. Microfibril‐associated glycoprotein‐1 binding to tropoelastin: multiple binding sites and the role of divalent cations. Eur J Biochem 2004271(14)3085–3090. [DOI] [PubMed] [Google Scholar]
- 90.Hanssen E, Hew F H, Moore E, Gibson M A. MAGP‐2 has multiple binding regions on fibrillins and has covalent periodic association with fibrillin‐containing microfibrils. J Biol Chem 2004279(28)29185–29194. [DOI] [PubMed] [Google Scholar]
- 91.Werneck C C, Trask B C, Broekelmann T J, Trask T M, Ritty T M, Segade F, Mecham R P. Identification of a major microfibril‐associated glycoprotein‐1‐binding domain in fibrillin‐2. J Biol Chem 2004279(22)23045–23051. [DOI] [PubMed] [Google Scholar]
- 92.Trask B C, Broekelmann T, Ritty T M, Trask T M, Tisdale C, Mecham R P. Posttranslational modifications of microfibril associated glycoprotein‐1 (MAGP‐1). Biochemistry 200140(14)4372–4380. [DOI] [PubMed] [Google Scholar]
- 93.Trask B C, Trask T M, Broekelmann T, Mecham R P. The microfibrillar proteins MAGP‐1 and fibrillin‐1 form a ternary complex with the chondroitin sulfate proteoglycan decorin. Mol Biol Cell 200011(5)1499–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gibson M A, Leavesley D I, Ashman L K. Microfibril‐associated glycoprotein‐2 specifically interacts with a range of bovine and human cell types via alphaVbeta3 integrin. J Biol Chem 1999274(19)13060–13065. [DOI] [PubMed] [Google Scholar]
- 95.Hirose H, Ozsvath K J, Xia S, Tilson M D. Molecular cloning of the complementary DNA for an additional member of the family of aortic aneurysm antigenic proteins. J Vasc Surg 199726(2)313–318. [DOI] [PubMed] [Google Scholar]
- 96.Xia S, Ozsvath K, Hirose H, Tilson M D. Partial amino acid sequence of a novel 40‐kDa human aortic protein, with vitronectin‐like, fibrinogen‐like, and calcium binding domains: aortic aneurysm‐associated protein‐40 (AAAP‐40) [human MAGP‐3, proposed]. Biochem Biophys Res Commun 1996219(1)36–39. [DOI] [PubMed] [Google Scholar]
- 97.Yeh H, Chow M, Abrams W R, Fan J, Foster J, Mitchell H, Muenke M, Rosenbloom J. Structure of the human gene encoding the associated microfibrillar protein (MFAP1) and localization to chromosome 15q15–q21. Genomics 199423(2)443–449. [DOI] [PubMed] [Google Scholar]
- 98.Horrigan S K, Rich C B, Streeten B W, Li Z Y, Foster J A. Characterization of an associated microfibril protein through recombinant DNA techniques. J Biol Chem 1992267(14)10087–10095. [PubMed] [Google Scholar]
- 99.Abrams W R, Ma R I, Kucich U, Bashir M M, Decker S, Tsipouras P, McPherson J D, Wasmuth J J, Rosenbloom J. Molecular cloning of the microfibrillar protein MFAP3 and assignment of the gene to human chromosome 5q32–q33.2. Genomics 199526(1)47–54. [DOI] [PubMed] [Google Scholar]
- 100.Hirano E, Fujimoto N, Tajima S, Akiyama M, Ishibashi A, Kobayashi R, Okamoto K. Expression of 36‐kDa microfibril‐associated glycoprotein (MAGP‐36) in human keratinocytes and its localization in skin. J Dermatol Sci 200228(1)60–67. [DOI] [PubMed] [Google Scholar]
- 101.Toyoshima T, Yamashita K, Furuichi H, Shishibori T, Itano T, Kobayashi R. Ultrastructural distribution of 36‐kD microfibril‐associated glycoprotein (MAGP‐36) in human and bovine tissues. J Histochem Cytochem 199947(8)1049–1056. [DOI] [PubMed] [Google Scholar]
- 102.Isogai Z, Ono R N, Ushiro S, Keene D R, Chen Y, Mazzieri R, Charbonneau N L, Reinhardt D P, Rifkin D B, Sakai L Y. Latent transforming growth factor beta‐binding protein 1 interacts with fibrillin and is a microfibril‐associated protein. J Biol Chem 2003278(4)2750–2757. [DOI] [PubMed] [Google Scholar]
- 103.Gibson M A, Hatzinikolas G, Davis E C, Baker E, Sutherland G R, Mecham R P. Bovine latent transforming growth factor beta 1‐binding protein 2: molecular cloning, identification of tissue isoforms, and immunolocalization to elastin‐associated microfibrils. Mol Cell Biol 199515(12)6932–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Isogai Z, Aspberg A, Keene D R, Ono R N, Reinhardt D P, Sakai L Y. Versican interacts with fibrillin‐1 and links extracellular microfibrils to other connective tissue networks. J Biol Chem 2002277(6)4565–4572. [DOI] [PubMed] [Google Scholar]
- 105.Schaefer L, Mihalik D, Babelova A, Krzyzankova M, Grone H J, Iozzo R V, Young M F, Seidler D G, Lin G, Reinhardt D P, Schaefer R M. Regulation of fibrillin‐1 by biglycan and decorin is important for tissue preservation in the kidney during pressure‐induced injury. Am J Pathol 2004165(2)383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Reinhardt D P, Sasaki T, Dzamba B J, Keene D R, Chu M L, Gohring W, Timpl R, Sakai L Y. Fibrillin‐1 and fibulin‐2 interact and are colocalized in some tissues. J Biol Chem 1996271(32)19489–19496. [DOI] [PubMed] [Google Scholar]
- 107.Freeman L J, Lomas A, Hodson N, Sherratt M J, Mellody K T, Weiss A S, Shuttleworth A, Kielty C M. Fibulin‐5 interacts with fibrillin‐1 molecules and microfibrils. Biochem J 20053881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bressan G M, Daga‐Gordini D, Colombatti A, Castellani I, Marigo V, Volpin D. Emilin, a component of elastic fibers preferentially located at the elastin‐microfibrils interface. J Cell Biol 1993121(1)201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dietz H C, Cutting C R, Pyeritz R E, Maslen C L, Sakai L Y, Corson G M, Puffenberger E G, Hamosh A, Nanthakumar E J, Curristin S M.et al Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature 1991352(6333)337–339. [DOI] [PubMed] [Google Scholar]
- 110.Putnam E A, Zhang H, Ramirez F, Milewicz D M. Fibrillin‐2 (FBN2) mutations result in the Marfan‐like disorder, congenital contractural arachnodactyly. Nat Genet 199511(4)456–458. [DOI] [PubMed] [Google Scholar]
- 111.Mizuguchi T, Collod‐Beroud G, Akiyama T, Abifadel M, Harada N, Morisaki T, Allard D, Varret M, Claustres M, Morisaki H, Ihara M, Kinoshita A, Yoshiura K, Junien C, Kajii T, Jondeau G, Ohta T, Kishino T, Furukawa Y, Nakamura Y, Niikawa N, Boileau C, Matsumoto N. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat Genet 200436(8)855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Loeys B L, Chen J, Neptune E R, Judge D P, Podowski M, Holm T, Meyers J, Leitch C C, Katsanis N, Sharifi N, Xu F L, Myers L A, Spevak P J, Cameron D E, De Backer J, Hellemans J, Chen Y, Davis E C, Webb C L, Kress W, Coucke P, Rifkin D B, De Paepe A M, Dietz H C. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet 200537(3)275–281. [DOI] [PubMed] [Google Scholar]
- 113.Pannu H, Fadulu V T, Chang J, Lafont A, Hasham S N, Sparks E, Giampietro P F, Zaleski C, Estrera A L, Safi H J, Shete S, Willing M C, Raman C S, Milewicz D M. Mutations in transforming growth factor‐beta receptor type II cause familial thoracic aortic aneurysms and dissections. Circulation 2005112(4)513–520. [DOI] [PubMed] [Google Scholar]
- 114.De Paepe A, Devereux R B, Dietz H C, Hennekam R C, Pyeritz R E. Revised diagnostic criteria for the Marfan syndrome. Am J Med Genet 199662(4)417–426. [DOI] [PubMed] [Google Scholar]
- 115.Pereira L, Levran O, Ramirez F, Lynch J R, Sykes B, Pyeritz R E, Dietz H C. A molecular approach to the stratification of cardiovascular risk in families with Marfan's syndrome. N Engl J Med 1994331(3)148–153. [DOI] [PubMed] [Google Scholar]
- 116.Tynan K, Comeau K, Pearson M, Wilgenbus P, Levitt D, Gasner C, Berg M A, Miller D C, Francke U. Mutation screening of complete fibrillin‐1 coding sequence: report of five new mutations, including two in 8‐cysteine domains. Hum Mol Genet 19932(11)1813–1821. [DOI] [PubMed] [Google Scholar]
- 117.Kainulainen K, Karttunen L, Puhakka L, Sakai L, Peltonen L. Mutations in the fibrillin gene responsible for dominant ectopia lentis and neonatal Marfan syndrome. Nat Genet 19946(1)64–69. [DOI] [PubMed] [Google Scholar]
- 118.Hayward C, Porteous M E, Brock D J. Mutation screening of all 65 exons of the fibrillin‐1 gene in 60 patients with Marfan syndrome: report of 12 novel mutations. Hum Mutat 199710(4)280–289. [DOI] [PubMed] [Google Scholar]
- 119.Nijbroek G, Sood S, McIntosh I, Francomano C A, Bull E, Pereira L, Ramirez F, Pyeritz R E, Dietz H C. Fifteen novel FBN1 mutations causing Marfan syndrome detected by heteroduplex analysis of genomic amplicons. Am J Hum Genet 199557(1)8–21. [PMC free article] [PubMed] [Google Scholar]
- 120.Katzke S, Booms P, Tiecke F, Palz M, Pletschacher A, Turkmen S, Neumann L M, Pregla R, Leitner C, Schramm C, Lorenz P, Hagemeier C, Fuchs J, Skovby F, Rosenberg T, Robinson P N. TGGE screening of the entire FBN1 coding sequence in 126 individuals with marfan syndrome and related fibrillinopathies. Hum Mutat 200220(3)197–208. [DOI] [PubMed] [Google Scholar]
- 121.Pepe G, Giusti B, Evangelisti L, Porciani M C, Brunelli T, Giurlani L, Attanasio M, Fattori R, Bagni C, Comeglio P, Abbate R, Gensini G F. Fibrillin‐1 (FBN1) gene frameshift mutations in Marfan patients: genotype‐phenotype correlation. Clin Genet 200159(6)444–450. [DOI] [PubMed] [Google Scholar]
- 122.Korkko J, Kaitila I, Lonnqvist L, Peltonen L, Ala‐Kokko L. Sensitivity of conformation sensitive gel electrophoresis in detecting mutations in Marfan syndrome and related conditions. J Med Genet 200239(1)34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Halliday D J, Hutchinson S, Lonie L, Hurst J A, Firth H, Handford P A, Wordsworth P. Twelve novel FBN1 mutations in Marfan syndrome and Marfan related phenotypes test the feasibility of FBN1 mutation testing in clinical practice. J Med Genet 200239(8)589–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Matyas G, De Paepe A, Halliday D, Boileau C, Pals G, Steinmann B. Evaluation and application of denaturing HPLC for mutation detection in Marfan syndrome: identification of 20 novel mutations and two novel polymorphisms in the FBN1 gene. Hum Mutat 200219(4)443–456. [DOI] [PubMed] [Google Scholar]
- 125.Loeys B, Nuytinck L, Delvaux I, De Bie S, De Paepe A. Genotype and phenotype analysis of 171 patients referred for molecular study of the fibrillin‐1 gene FBN1 because of suspected Marfan syndrome. Arch Intern Med 2001161(20)2447–2454. [DOI] [PubMed] [Google Scholar]
- 126.Rommel K, Karck M, Haverich A, Schmidtke J, Arslan‐Kirchner M. Mutation screening of the fibrillin‐1 (FBN1) gene in 76 unrelated patients with Marfan syndrome or Marfanoid features leads to the identification of 11 novel and three previously reported mutations. Hum Mutat 200220(5)406–407. [DOI] [PubMed] [Google Scholar]
- 127.Schrijver I, Liu W, Odom R, Brenn T, Oefner P, Furthmayr H, Francke U. Premature termination mutations in FBN1: distinct effects on differential allelic expression and on protein and clinical phenotypes. Am J Hum Genet 200271(2)223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Loeys B, De Backer J, Van Acker P, Wettinck K, Pals G, Nuytinck L, Coucke P, De Paepe A. Comprehensive molecular screening of the FBN1 gene favors locus homogeneity of classical Marfan syndrome. Hum Mutat 200424(2)140–146. [DOI] [PubMed] [Google Scholar]
- 129.Booms P, Cisler J, Mathews K R, Godfrey M, Tiecke F, Kaufmann U C, Vetter U, Hagemeier C, Robinson P N. Novel exon skipping mutation in the fibrillin‐1 gene: two ‘hot spots' for the neonatal Marfan syndrome. Clin Genet 199955(2)110–117. [DOI] [PubMed] [Google Scholar]
- 130.Putnam E A, Cho M, Zinn A B, Towbin J A, Byers P H, Milewicz D M. Delineation of the Marfan phenotype associated with mutations in exons 23–32 of the FBN1 gene. Am J Med Genet 199662(3)233–242. [DOI] [PubMed] [Google Scholar]
- 131.Tiecke F, Katzke S, Booms P, Robinson P N, Neumann L, Godfrey M, Mathews K R, Scheuner M, Hinkel G K, Brenner R E, Hovels‐Gurich H H, Hagemeier C, Fuchs J, Skovby F, Rosenberg T. Classic, atypically severe and neonatal Marfan syndrome: twelve mutations and genotype‐phenotype correlations in FBN1 exons 24–40. Eur J Hum Genet 20019(1)13–21. [DOI] [PubMed] [Google Scholar]
- 132.Lönnqvist L, Child A, Kainulainen K, Davidson R, Puhakka L, Peltonen L. A novel mutation of the fibrillin gene causing ectopia lentis. Genomics 199419(3)573–576. [DOI] [PubMed] [Google Scholar]
- 133.Ades L C, Holman K J, Brett M S, Edwards M J, Bennetts B. Ectopia lentis phenotypes and the FBN1 gene. Am J Med Genet A 2004126(3)284–289. [DOI] [PubMed] [Google Scholar]
- 134.Ades L C, Sreetharan D, Onikul E, Stockton V, Watson K C, Holman K J. Segregation of a novel FBN1 gene mutation, G1796E, with kyphoscoliosis and radiographic evidence of vertebral dysplasia in three generations. Am J Med Genet 2002109(4)261–270. [DOI] [PubMed] [Google Scholar]
- 135.Hayward C, Porteous M E, Brock D J. A novel mutation in the fibrillin gene (FBN1) in familial arachnodactyly. Mol Cell Probes 19948(4)325–327. [DOI] [PubMed] [Google Scholar]
- 136.Dietz H C, McIntosh I, Sakai L Y, Corson G M, Chalberg S C, Pyeritz R E, Francomano C A. Four novel FBN1 mutations: significance for mutant transcript level and EGF‐like domain calcium binding in the pathogenesis of Marfan syndrome. Genomics 199317(2)468–475. [DOI] [PubMed] [Google Scholar]
- 137.Sood S, Eldadah Z A, Krause W L, McIntosh I, Dietz H C. Mutation in fibrillin‐1 and the Marfanoid‐craniosynostosis (Shprintzen‐Goldberg) syndrome. Nat Genet 199612(2)209–211. [DOI] [PubMed] [Google Scholar]
- 138.Kosaki K, Takahashi D, Udaka T, Kosaki R, Matsumoto M, Ibe S, Isobe T, Tanaka Y, Takahashi T. Molecular pathology of Shprintzen‐Goldberg syndrome. Am J Med Genet A 2006140A(1)104–108. [DOI] [PubMed] [Google Scholar]
- 139.Robinson P N, Neumann L M, Tinschert S. Response to Kosaki et al. “Molecular pathology of Shprintzen‐Goldberg syndrome”. Am J Med Genet A 2006140A(1)109–110. [DOI] [PubMed] [Google Scholar]
- 140.Ståhl‐Hallengren C, Ukkonen T, Kainulainen K, Kristofersson U, Saxne T, Tornqvist K, Peltonen L. An extra cysteine in one of the non‐calcium‐binding epidermal growth factor‐like motifs of the FBN1 polypeptide is connected to a novel variant of Marfan syndrome. J Clin Invest 199494(2)709–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Black C, Withers A P, Gray J R, Bridges A B, Craig A, Baty D U, Boxer M. Correlation of a recurrent FBN1 mutation (R122C) with an atypical familial Marfan syndrome phenotype. Hum Mutat 1998Suppl 1S198–S200. [DOI] [PubMed]
- 142.Faivre L, Gorlin R J, Wirtz M K, Godfrey M, Dagoneau N, Samples J R, Le Merrer M, Collod‐Beroud G, Boileau C, Munnich A, Cormier‐Daire V. In frame fibrillin‐1 gene deletion in autosomal dominant Weill‐Marchesani syndrome. J Med Genet 200340(1)34–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cariello N F, Cui L, Beroud C, Soussi T. Database and software for the analysis of mutations in the human p53 gene. Cancer Res 199454(16)4454–4460. [PubMed] [Google Scholar]
- 144.Beroud C, Collod‐Beroud G, Boileau C, Soussi T, Junien C. UMD (universal mutation database): a generic software to build and analyze locus‐specific databases. Hum Mutat 200015(1)86–94. [DOI] [PubMed] [Google Scholar]
- 145.Beroud C, Hamroun D, Collod‐Beroud G, Boileau C, Soussi T, Claustres M. UMD (Universal Mutation Database): 2005 update. Hum Mutat 200526(3)184–191. [DOI] [PubMed] [Google Scholar]
- 146.Collod G, Beroud C, Soussi T, Junien C, Boileau C. Software and database for the analysis of mutations in the human FBN1 gene. Nucleic Acids Res 199624(1)137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Collod‐Beroud G, Beroud C, Ades L, Black C, Boxer M, Brock D J, Godfrey M, Hayward C, Karttunen L, Milewicz D, Peltonen L, Richards R I, Wang M, Junien C, Boileau C. Marfan database (second edition): software and database for the analysis of mutations in the human FBN1 gene. Nucleic Acids Res 199725(1)147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Collod‐Beroud G, Beroud C, Ades L, Black C, Boxer M, Brock D J, Holman K J, de Paepe A, Francke U, Grau U, Hayward C, Klein H G, Liu W, Nuytinck L, Peltonen L, Alvarez Perez A B, Rantamaki T, Junien C, Boileau C. Marfan database (third edition): new mutations and new routines for the software. Nucleic Acids Res 199826(1)229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Collod‐Beroud G, Le Bourdelles S, Ades L, Ala‐Kokko L, Booms P, Boxer M, Child A, Comeglio P, De Paepe A, Hyland J C, Holman K, Kaitila I, Loeys B, Matyas G, Nuytinck L, Peltonen L, Rantamaki T, Robinson P, Steinmann B, Junien C, Beroud C, Boileau C. Update of the UMD‐FBN1 mutation database and creation of an FBN1 polymorphism database. Hum Mutat 200322(3)199–208. [DOI] [PubMed] [Google Scholar]
- 150.den Dunnen J T, Antonarakis S E. Nomenclature for the description of human sequence variations. Hum Genet 2001109(1)121–124. [DOI] [PubMed] [Google Scholar]
- 151.Liu W, Qian C, Comeau K, Brenn T, Furthmayr H, Francke U. Mutant fibrillin‐1 monomers lacking EGF‐like domains disrupt microfibril assembly and cause severe marfan syndrome. Hum Mol Genet 19965(10)1581–1587. [DOI] [PubMed] [Google Scholar]
- 152.Guo D, Tan F K, Cantu A, Plon S E, Milewicz D M. FBN1 exon 2 splicing error in a patient with Marfan syndrome. Am J Med Genet 2001101(2)130–134. [DOI] [PubMed] [Google Scholar]
- 153.Frischmeyer P A, Dietz H C. Nonsense‐mediated mRNA decay in health and disease. Hum Mol Genet 19998(10)1893–1900. [DOI] [PubMed] [Google Scholar]
- 154.Liu W, Qian C, Francke U. Silent mutation induces exon skipping of fibrillin‐1 gene in Marfan syndrome. Nat Genet 199716(4)328–329. [DOI] [PubMed] [Google Scholar]
- 155.Dietz H C, Kendzior R J., Jr Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat Genet 19948(2)183–188. [DOI] [PubMed] [Google Scholar]
- 156.Caputi M, Kendzior R J, Jr, Beemon K L. A nonsense mutation in the fibrillin‐1 gene of a Marfan syndrome patient induces NMD and disrupts an exonic splicing enhancer. Genes Dev 200216(14)1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Collod‐Beroud G, Boileau C. Marfan syndrome in the third millennium. Eur J Hum Genet 200210(11)673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Palz M, Tiecke F, Booms P, Goldner B, Rosenberg T, Fuchs J, Skovby F, Schumacher H, Kaufmann U C, von Kodolitsch Y, Nienaber C A, Leitner C, Katzke S, Vetter B, Hagemeier C, Robinson P N. Clustering of mutations associated with mild Marfan‐like phenotypes in the 3′ region of FBN1 suggests a potential genotype‐phenotype correlation. Am J Med Genet 200091(3)212–221. [DOI] [PubMed] [Google Scholar]
- 159.Hutchinson S, Furger A, Halliday D, Judge D P, Jefferson A, Dietz H C, Firth H, Handford P A. Allelic variation in normal human FBN1 expression in a family with Marfan syndrome: a potential modifier of phenotype? Hum Mol Genet 200312(18)2269–2276. [DOI] [PubMed] [Google Scholar]
- 160.Giusti B, Marcucci R, Lapini I, Sestini I, Lenti M, Yacoub M, Pepe G. Role of hyperhomocysteinemia in aortic disease. Cell Mol Biol (Noisy‐le‐grand) 200450(8)945–952. [PubMed] [Google Scholar]
- 161.Shi Y, Massague J. Mechanisms of TGF‐beta signaling from cell membrane to the nucleus. Cell 2003113(6)685–700. [DOI] [PubMed] [Google Scholar]
- 162.Boileau C, Jondeau G, Babron M C, Coulon M, Alexandre J A, Sakai L, Melki J, Delorme G, Dubourg O, Bonaiti‐Pellie C.et al Autosomal dominant Marfan‐like connective‐tissue disorder with aortic dilation and skeletal anomalies not linked to the fibrillin genes. Am J Hum Genet 199353(1)46–54. [PMC free article] [PubMed] [Google Scholar]
- 163.Collod G, Babron M C, Jondeau G, Coulon M, Weissenbach J, Dubourg O, Bourdarias J P, Bonaiti‐Pellie C, Junien C, Boileau C. A second locus for Marfan syndrome maps to chromosome 3p24.2–p25. Nat Genet 19948(3)264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Parsons R, Myeroff L L, Liu B, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Microsatellite instability and mutations of the transforming growth factor beta type II receptor gene in colorectal cancer. Cancer Res 199555(23)5548–5550. [PubMed] [Google Scholar]
- 165.Shin K H, Park Y J, Park J G. Mutational analysis of the transforming growth factor beta receptor type II gene in hereditary nonpolyposis colorectal cancer and early‐onset colorectal cancer patients. Clin Cancer Res 20006(2)536–540. [PubMed] [Google Scholar]
- 166.Markowitz S. TGF‐beta receptors and DNA repair genes, coupled targets in a pathway of human colon carcinogenesis. Biochim Biophys Acta 20001470(1)M13–M20. [DOI] [PubMed] [Google Scholar]
- 167.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B.et al Inactivation of the type II TGF‐beta receptor in colon cancer cells with microsatellite instability. Science 1995268(5215)1336–1338. [DOI] [PubMed] [Google Scholar]
- 168.Disabella E, Grasso M, Marziliano N, Ansaldi S, Lucchelli C, Porcu E, Tagliani M, Pilotto A, Diegoli M, Lanzarini L, Malattia C, Pelliccia A, Ficcadenti A, Gabrielli O, Arbustini E. Two novel and one known mutation of the TGFBR2 gene in Marfan syndrome not associated with FBN1 gene defects. Eur J Hum Genet 200614(1)34–38. [DOI] [PubMed] [Google Scholar]
- 169.Hecht F, Beals R K. “New” syndrome of congenital contractural arachnodactyly originally described by Marfan in 1896. Pediatrics 197249(4)574–579. [PubMed] [Google Scholar]
- 170.Epstein C J, Graham C B, Hodgkin W E, Hecht F, Motulsky A G. Hereditary dysplasia of bone with kyphoscoliosis, contractures, and abnormally shaped ears. J Pediatr 196873(3)379–386. [DOI] [PubMed] [Google Scholar]
- 171.Ramos Arroyo M A, Weaver D D, Beals R K. Congenital contractural arachnodactyly. Report of four additional families and review of literature. Clin Genet 198527(6)570–581. [DOI] [PubMed] [Google Scholar]
- 172.Viljoen D. Congenital contractural arachnodactyly (Beals syndrome). J Med Genet 199431(8)640–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jones J L, Lane J E, Logan J J, Vanegas M E. Beals‐Hecht syndrome. South Med J 200295(7)753–755. [PubMed] [Google Scholar]
- 174.Gupta P A, Putnam E A, Carmical S G, Kaitila I, Steinmann B, Child A, Danesino C, Metcalfe K, Berry S A, Chen E, Delorme C V, Thong M K, Ades L C, Milewicz D M. Ten novel FBN2 mutations in congenital contractural arachnodactyly: delineation of the molecular pathogenesis and clinical phenotype. Hum Mutat 200219(1)39–48. [DOI] [PubMed] [Google Scholar]
- 175.Park E S, Putnam E A, Chitayat D, Child A, Milewicz D M. Clustering of FBN2 mutations in patients with congenital contractural arachnodactyly indicates an important role of the domains encoded by exons 24 through 34 during human development. Am J Med Genet 199878(4)350–355. [PubMed] [Google Scholar]
- 176.Belleh S, Zhou G, Wang M, Der Kaloustian V M, Pagon R A, Godfrey M. Two novel fibrillin‐2 mutations in congenital contractural arachnodactyly. Am J Med Genet 200092(1)7–12. [PubMed] [Google Scholar]
- 177.Maslen C, Babcock D, Raghunath M, Steinmann B. A rare branch‐point mutation is associated with missplicing of fibrillin‐2 in a large family with congenital contractural arachnodactyly. Am J Hum Genet 199760(6)1389–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Babcock D, Gasner C, Francke U, Maslen C. A single mutation that results in an Asp to His substitution and partial exon skipping in a family with congenital contractural arachnodactyly. Hum Genet 1998103(1)22–28. [DOI] [PubMed] [Google Scholar]
- 179.Wang M, Clericuzio C L, Godfrey M. Familial occurrence of typical and severe lethal congenital contractural arachnodactyly caused by missplicing of exon 34 of fibrillin‐2. Am J Hum Genet 199659(5)1027–1034. [PMC free article] [PubMed] [Google Scholar]
- 180.Gupta P A, Wallis D D, Chin T O, Northrup H, Tran‐Fadulu V T, Towbin J A, Milewicz D M. FBN2 mutation associated with manifestations of Marfan syndrome and congenital contractural arachnodactyly. J Med Genet 200441(5)e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Quondamatteo F, Reinhardt D P, Charbonneau N L, Pophal G, Sakai L Y, Herken R. Fibrillin‐1 and fibrillin‐2 in human embryonic and early fetal development. Matrix Biol 200221(8)637–646. [DOI] [PubMed] [Google Scholar]
- 182.Mariencheck M C, Davis E C, Zhang H, Ramirez F, Rosenbloom J, Gibson M A, Parks W C, Mecham R P. Fibrillin‐1 and fibrillin‐2 show temporal and tissue‐specific regulation of expression in developing elastic tissues. Connect Tissue Res 199531(2)87–97. [DOI] [PubMed] [Google Scholar]
- 183.Wunsch A M, Little C D, Markwald R R. Cardiac endothelial heterogeneity defines valvular development as demonstrated by the diverse expression of JB3, an antigen of the endocardial cushion tissue. Dev Biol 1994165(2)585–601. [DOI] [PubMed] [Google Scholar]
- 184.Rongish B J, Drake C J, Argraves W S, Little C D. Identification of the developmental marker, JB3‐antigen, as fibrillin‐2 and its de novo organization into embryonic microfibrous arrays. Dev Dyn 1998212(3)461–471. [DOI] [PubMed] [Google Scholar]
- 185.Yang Q, Ota K, Tian Y, Kumar A, Wada J, Kashihara N, Wallner E, Kanwar Y S. Cloning of rat fibrillin‐2 cDNA and its role in branching morphogenesis of embryonic lung. Dev Biol 1999212(1)229–242. [DOI] [PubMed] [Google Scholar]
- 186.Browning V L, Chaudhry S S, Planchart A, Dixon M J, Schimenti J C. Mutations of the mouse Twist and sy (fibrillin 2) genes induced by chemical mutagenesis of ES cells. Genomics 200173(3)291–298. [DOI] [PubMed] [Google Scholar]
- 187.Johnson K R, Cook S A, Zheng Q Y. The original shaker‐with‐syndactylism mutation (sy) is a contiguous gene deletion syndrome. Mamm Genome 19989(11)889–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Chaudhry S S, Gazzard J, Baldock C, Dixon J, Rock M J, Skinner G C, Steel K P, Kielty C M, Dixon M J. Mutation of the gene encoding fibrillin‐2 results in syndactyly in mice. Hum Mol Genet 200110(8)835–843. [DOI] [PubMed] [Google Scholar]
- 189.Milewicz D M, Chen H, Park E S, Petty E M, Zaghi H, Shashidhar G, Willing M, Patel V. Reduced penetrance and variable expressivity of familial thoracic aortic aneurysms/dissections. Am J Cardiol 199882(4)474–479. [DOI] [PubMed] [Google Scholar]
- 190.Nicod P, Bloor C, Godfrey M, Hollister D, Pyeritz R E, Dittrich H, Polikar R, Peterson K L. Familial aortic dissecting aneurysm. J Am Coll Cardiol 198913(4)811–819. [DOI] [PubMed] [Google Scholar]
- 191.Lie J T. Aortic dissection in Turner's syndrome. Am Heart J 1982103(6)1077–1080. [DOI] [PubMed] [Google Scholar]
- 192.Shachter N, Perloff J K, Mulder D G. Aortic dissection in Noonan's syndrome (46 XY turner). Am J Cardiol 198454(3)464–465. [DOI] [PubMed] [Google Scholar]
- 193.Leier C V, Call T D, Fulkerson P K, Wooley C F. The spectrum of cardiac defects in the Ehlers‐Danlos syndrome, types I and III. Ann Intern Med 198092(2 Pt 1)171–178. [DOI] [PubMed] [Google Scholar]
- 194.Lund J T, Jensen M B, Hjelms E. Aneurysm of the ductus arteriosus. A review of the literature and the surgical implications. Eur J Cardiothorac Surg 19915(11)566–570. [DOI] [PubMed] [Google Scholar]
- 195.Cusick D A, Frederiksen J W, Mehlman D J. Acute aortic dissection: association with patent ductus arteriosus. Am J Card Imaging 199610(3)200–203. [PubMed] [Google Scholar]
- 196.McKusick V A, Logue R B, Bahson H T. Association of aortic valvular disease and cystic medial necrosis of the ascending aorta. Circulation 195716188–190. [DOI] [PubMed] [Google Scholar]
- 197.McKusick V A. Association of congenital bicuspid aortic valve and Erdheim's cystic medial necrosis. Lancet 19721(7758)1026–1027. [DOI] [PubMed] [Google Scholar]
- 198.Biddinger A, Rocklin M, Coselli J, Milewicz D M. Familial thoracic aortic dilatations and dissections: a case control study. J Vasc Surg 199725(3)506–511. [DOI] [PubMed] [Google Scholar]
- 199.Coady M A, Rizzo J A, Goldstein L J, Elefteriades J A. Natural history, pathogenesis, and etiology of thoracic aortic aneurysms and dissections. Cardiol Clin 199917(4)615–35 vii. [DOI] [PubMed] [Google Scholar]
- 200.Francke U, Berg M A, Tynan K, Brenn T, Liu W, Aoyama T, Gasner C, Miller D C, Furthmayr H. A Gly1127Ser mutation in an EGF‐like domain of the fibrillin‐1 gene is a risk factor for ascending aortic aneurysm and dissection. Am J Hum Genet 199556(6)1287–1296. [PMC free article] [PubMed] [Google Scholar]
- 201.Tromp G, Wu Y, Prockop D J, Madhatheri S L, Kleinert C, Earley J J, Zhuang J, Norrgard O, Darling R C, Abbott W M.et al Sequencing of cDNA from 50 unrelated patients reveals that mutations in the triple‐helical domain of type III procollagen are an infrequent cause of aortic aneurysms. J Clin Invest 199391(6)2539–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Guo D, Hasham S, Kuang S Q, Vaughan C J, Boerwinkle E, Chen H, Abuelo D, Dietz H C, Basson C T, Shete S S, Milewicz D M. Familial thoracic aortic aneurysms and dissections: genetic heterogeneity with a major locus mapping to 5q13–14. Circulation 2001103(20)2461–2468. [DOI] [PubMed] [Google Scholar]
- 203.Kakko S, Raisanen T, Tamminen M, Airaksinen J, Groundstroem K, Juvonen T, Ylitalo A, Uusimaa P, Savolainen M J. Candidate locus analysis of familial ascending aortic aneurysms and dissections confirms the linkage to the chromosome 5q13–14 in Finnish families. J Thorac Cardiovasc Surg 2003126(1)106–113. [DOI] [PubMed] [Google Scholar]
- 204.Vaughan C J, Casey M, He J, Veugelers M, Henderson K, Guo D, Campagna R, Roman M J, Milewicz D M, Devereux R B, Basson C T. Identification of a chromosome 11q23.2–q24 locus for familial aortic aneurysm disease, a genetically heterogeneous disorder. Circulation 2001103(20)2469–2475. [DOI] [PubMed] [Google Scholar]
- 205.Hasham S N, Willing M C, Guo D C, Muilenburg A, He R, Tran V T, Scherer S E, Shete S S, Milewicz D M. Mapping a locus for familial thoracic aortic aneurysms and dissections (TAAD2) to 3p24–25. Circulation 2003107(25)3184–3190. [DOI] [PubMed] [Google Scholar]
- 206.Smallridge R S, Whiteman P, Werner J M, Campbell I D, Handford P A, Downing A K. Solution structure and dynamics of a calcium binding epidermal growth factor‐like domain pair from the neonatal region of human fibrillin‐1. J Biol Chem 2003278(14)12199–12206. [DOI] [PubMed] [Google Scholar]
- 207.Smallridge R S, Whiteman P, Doering K, Handford P A, Downing A K. EGF‐like domain calcium affinity modulated by N‐terminal domain linkage in human fibrillin‐1. J Mol Biol 1999286(3)661–668. [DOI] [PubMed] [Google Scholar]
- 208.Werner J M, Knott V, Handford P A, Campbell I D, Downing A K. Backbone dynamics of a cbEGF domain pair in the presence of calcium. J Mol Biol 2000296(4)1065–1078. [DOI] [PubMed] [Google Scholar]
- 209.Reinhardt D P, Ono R N, Sakai L Y. Calcium stabilizes fibrillin‐1 against proteolytic degradation. J Biol Chem 1997272(2)1231–1236. [DOI] [PubMed] [Google Scholar]
- 210.McGettrick A J, Knott V, Willis A, Handford P A. Molecular effects of calcium binding mutations in Marfan syndrome depend on domain context. Hum Mol Genet 20009(13)1987–1994. [DOI] [PubMed] [Google Scholar]
- 211.Knott V, Downing A K, Cardy C M, Handford P. Calcium binding properties of an epidermal growth factor‐like domain pair from human fibrillin‐1. J Mol Biol 1996255(1)22–27. [DOI] [PubMed] [Google Scholar]
- 212.Suk J Y, Jensen S, McGettrick A, Willis A C, Whiteman P, Redfield C, Handford P A. Structural consequences of cysteine substitutions C1977Y and C1977R in calcium‐binding epidermal growth factor‐like domain 30 of human fibrillin‐1. J Biol Chem 2004279(49)51258–51265. [DOI] [PubMed] [Google Scholar]
- 213.Kettle S, Yuan X, Grundy G, Knott V, Downing A K, Handford P A. Defective calcium binding to fibrillin‐1: consequence of an N2144S change for fibrillin‐1 structure and function. J Mol Biol 1999285(3)1277–1287. [DOI] [PubMed] [Google Scholar]
- 214.Vollbrandt T, Tiedemann K, El‐Hallous E, Lin G, Brinckmann J, John H, Bätge B, Notbohm H, Reinhardt D P. Consequences of cysteine mutations in calcium‐binding epidermal growth factor modules of fibrillin‐1. J Biol Chem 2004279(31)32924–32931. [DOI] [PubMed] [Google Scholar]
- 215.Whiteman P, Downing A K, Smallridge R, Winship P R, Handford P A. A Gly —>Ser change causes defective folding in vitro of calcium‐binding epidermal growth factor‐like domains from factor IX and fibrillin‐1. J Biol Chem 1998273(14)7807–7813. [DOI] [PubMed] [Google Scholar]
- 216.Whiteman P, Smallridge R S, Knott V, Cordle J J, Downing A K, Handford P A. A G1127S change in calcium‐binding epidermal growth factor‐like domain 13 of human fibrillin‐1 causes short range conformational effects. J Biol Chem 2001276(20)17156–17162. [DOI] [PubMed] [Google Scholar]
- 217.Reinhardt D P, Ono R N, Notbohm H, Muller P K, Bachinger H P, Sakai L Y. Mutations in calcium‐binding epidermal growth factor modules render fibrillin‐1 susceptible to proteolysis. A potential disease‐causing mechanism in Marfan syndrome. J Biol Chem 2000275(16)12339–12345. [DOI] [PubMed] [Google Scholar]
- 218.Schrijver I, Liu W, Brenn T, Furthmayr H, Francke U. Cysteine substitutions in epidermal growth factor‐like domains of fibrillin‐1: distinct effects on biochemical and clinical phenotypes. Am J Hum Genet 199965(4)1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Whiteman P, Handford P A. Defective secretion of recombinant fragments of fibrillin‐1: implications of protein misfolding for the pathogenesis of Marfan syndrome and related disorders. Hum Mol Genet 200312(7)727–737. [DOI] [PubMed] [Google Scholar]
- 220.Handford P, Downing A K, Rao Z, Hewett D R, Sykes B C, Kielty C M. The calcium binding properties and molecular organization of epidermal growth factor‐like domains in human fibrillin‐1. J Biol Chem 1995270(12)6751–6756. [DOI] [PubMed] [Google Scholar]
- 221.Whiteman P, Downing A K, Handford P A. NMR analysis of cbEGF domains gives new insights into the structural consequences of a P1148A substitution in fibrillin‐1. Protein Eng 199811(11)957–959. [DOI] [PubMed] [Google Scholar]
- 222.Kielty C M, Shuttleworth C A. The role of calcium in the organization of fibrillin microfibrils. FEBS Lett 1993336(2)323–326. [DOI] [PubMed] [Google Scholar]
- 223.Cardy C M, Handford P A. Metal ion dependency of microfibrils supports a rod‐like conformation for fibrillin‐1 calcium‐binding epidermal growth factor‐like domains. J Mol Biol 1998276(5)855–860. [DOI] [PubMed] [Google Scholar]
- 224.Wess T J, Purslow P P, Sherratt M J, Ashworth J, Shuttleworth C A, Kielty C M. Calcium determines the supramolecular organization of fibrillin‐rich microfibrils. J Cell Biol 1998141(3)829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sasaki T, Mann K, Murphy G, Chu M L, Timpl R. Different susceptibilities of fibulin‐1 and fibulin‐2 to cleavage by matrix metalloproteinases and other tissue proteases. Eur J Biochem 1996240(2)427–434. [DOI] [PubMed] [Google Scholar]
- 226.Colosetti P, Hellman U, Heldin C H, Miyazono K. Ca2+ binding of latent transforming growth factor‐beta 1 binding protein. FEBS Lett 1993320(2)140–144. [DOI] [PubMed] [Google Scholar]
- 227.Hyytiäinen M, Taipale J, Heldin C H, Keski‐Oja J. Recombinant latent transforming growth factor beta‐binding protein 2 assembles to fibroblast extracellular matrix and is susceptible to proteolytic processing and release. J Biol Chem 1998273(32)20669–20676. [DOI] [PubMed] [Google Scholar]
- 228.Booms P, Tiecke F, Rosenberg T, Hagemeier C, Robinson P N. Differential effect of FBN1 mutations on in vitro proteolysis of recombinant fibrillin‐1 fragments. Hum Genet 2000107(3)216–224. [DOI] [PubMed] [Google Scholar]
- 229.Robinson P N, Booms P. The molecular pathogenesis of the Marfan syndrome. Cell Mol Life Sci 200158(11)1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Fleischer K J, Nousari H C, Anhalt G J, Stone C D, Laschinger J C. Immunohistochemical abnormalities of fibrillin in cardiovascular tissues in Marfan's syndrome. Ann Thorac Surg 199763(4)1012–1017. [DOI] [PubMed] [Google Scholar]
- 231.Segura A M, Luna R E, Horiba K, Stetler‐Stevenson W G, McAllister H A, Jr, Willerson J T, Ferrans V J. Immunohistochemistry of matrix metalloproteinases and their inhibitors in thoracic aortic aneurysms and aortic valves of patients with Marfan's syndrome. Circulation 199898(19 Suppl)II331–7 discussion II337–8. [PubMed] [Google Scholar]
- 232.Sachdev N H, Di Girolamo N, McCluskey P J, Jennings A V, McGuinness R, Wakefield D, Coroneo M T. Lens dislocation in Marfan syndrome: potential role of matrix metalloproteinases in fibrillin degradation. Arch Ophthalmol 2002120(6)833–835. [PubMed] [Google Scholar]
- 233.Ashworth J L, Murphy G, Rock M J, Sherratt M J, Shapiro S D, Shuttleworth C A, Kielty C M. Fibrillin degradation by matrix metalloproteinases: implications for connective tissue remodelling. Biochem J 1999340(Pt 1)171–181. [PMC free article] [PubMed] [Google Scholar]
- 234.Hering T M. Regulation of chondrocyte gene expression. Front Biosci 19994D743–D761. [DOI] [PubMed] [Google Scholar]
- 235.Homandberg G A. Potential regulation of cartilage metabolism in osteoarthritis by fibronectin fragments. Front Biosci 19994D713–D730. [DOI] [PubMed] [Google Scholar]
- 236.Hu B, Kapila Y L, Buddhikot M, Shiga M, Kapila S. Coordinate induction of collagenase‐1, stromelysin‐1 and urokinase plasminogen activator (uPA) by the 120‐kDa cell‐binding fibronectin fragment in fibrocartilaginous cells: uPA contributes to activation of procollagenase‐1. Matrix Biol 200019(7)657–669. [DOI] [PubMed] [Google Scholar]
- 237.Loeser R F, Forsyth C B, Samarel A M, Im H J. Fibronectin‐fragment activation of proline‐rich tyrosine kinase PYK2 mediates integrin signals regulating collagenase‐3 expression by human chondrocytes through a protein kinase C‐dependent pathway. J Biol Chem 2003278(27)24577–24585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Booms P, Pregla R, Ney A, Barthel F, Reinhardt D P, Pletschacher A, Mundlos S, Robinson P N. RGD‐containing fibrillin‐1 fragments upregulate matrix metalloproteinase expression in cell culture: a potential factor in the pathogenesis of the Marfan syndrome. Hum Genet 2005116(1–2)51–61. [DOI] [PubMed] [Google Scholar]
- 239.Booms P, Ney A, Barthel F, Moroy G, Counsell D, Gille C, Guo G, Pregla R, Mundlos S, Alix A J, Robinson P N. A fibrillin‐1 fragment containing the elastin‐binding protein GxxPG consensus sequence upregulates matrix metalloproteinase‐1: biochemical and computational analysis. J Mol Cell Cardiol 200640(2)234–246. [DOI] [PubMed] [Google Scholar]
- 240.Munshi H G, Wu Y I, Mukhopadhyay S, Ottaviano A J, Sassano A, Koblinski J E, Platanias L C, Stack M S. Differential regulation of membrane type 1‐matrix metalloproteinase activity by ERK 1/2‐ and p38 MAPK‐modulated tissue inhibitor of metalloproteinases 2 expression controls transforming growth factor‐beta1‐induced pericellular collagenolysis. J Biol Chem 2004279(37)39042–39050. [DOI] [PubMed] [Google Scholar]
- 241.Bunton T E, Biery N J, Myers L, Gayraud B, Ramirez F, Dietz H C. Phenotypic alteration of vascular smooth muscle cells precedes elastolysis in a mouse model of Marfan syndrome. Circ Res 200188(1)37–43. [DOI] [PubMed] [Google Scholar]
- 242.Judge D P, Biery N J, Keene D R, Geubtner J, Myers L, Huso D L, Sakai L Y, Dietz H C. Evidence for a critical contribution of haploinsufficiency in the complex pathogenesis of Marfan syndrome. J Clin Invest 2004114(2)172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Neptune E R, Frischmeyer P A, Arking D E, Myers L, Bunton T E, Gayraud B, Ramirez F, Sakai L Y, Dietz H C. Dysregulation of TGF‐beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet 200333(3)407–411. [DOI] [PubMed] [Google Scholar]
- 244.Ng C M, Cheng A, Myers L A, Martinez‐Murillo F, Jie C, Bedja D, Gabrielson K L, Hausladen J M, Mecham R P, Judge D P, Dietz H C. TGF‐β‐dependent pathogenesis of mitral valve prolapse in a mouse model of Marfan syndrome. J Clin Invest 2004114(11)1586–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ramirez F, Rifkin D B. Cell signaling events: a view from the matrix. Matrix Biol 200322(2)101–107. [DOI] [PubMed] [Google Scholar]
- 246.Wood J R, Bellamy D, Child A H, Citron K M. Pulmonary disease in patients with Marfan syndrome. Thorax 198439(10)780–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Rigante D, Segni G, Bush A. Persistent spontaneous pneumothorax in an adolescent with Marfan's syndrome and pulmonary bullous dysplasia. Respiration 200168(6)621–624. [DOI] [PubMed] [Google Scholar]
- 248.Dietz H C, Loeys B, Carta L, Ramirez F. Recent progress towards a molecular understanding of Marfan syndrome. Am J Med Genet C Semin Med Genet 2005139(1)4–9. [DOI] [PubMed] [Google Scholar]
- 249.Corson G M, Chalberg S C, Dietz H C, Charbonneau N L, Sakai L Y. Fibrillin binds calcium and is coded by cDNAs that reveal a multidomain structure and alternatively spliced exons at the 5′ end. Genomics 199317(2)476–484. [DOI] [PubMed] [Google Scholar]
- 250.Grady W M, Rajput A, Myeroff L, Liu D F, Kwon K, Willis J, Markowitz S. Mutation of the type II transforming growth factor‐beta receptor is coincident with the transformation of human colon adenomas to malignant carcinomas. Cancer Res 199858(14)3101–3104. [PubMed] [Google Scholar]
- 251.Lücke C D, Philpott A, Metcalfe J C, Thompson A M, Hughes‐Davies L, Kemp P R, Hesketh R. Inhibiting mutations in the transforming growth factor beta type 2 receptor in recurrent human breast cancer. Cancer Res 200161(2)482–485. [PubMed] [Google Scholar]