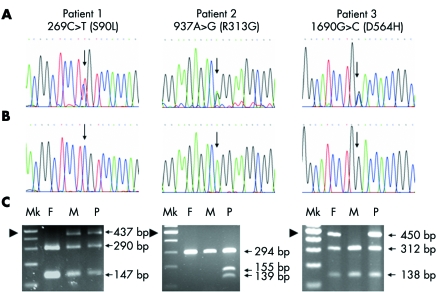
Figure 2 Mutation analysis. The left, middle, and right panels relate to patients 1, 2, and 3, respectively. Electropherograms of (A) patients, showing 269C→T, 937A→G, and 1690G→C (arrows) in patients 1, 2, and 3, respectively; and (B) controls, showing normal genotypes at codons 269 as CC, 937 AA and 1690 GG (arrows). (C) Restriction enzyme digestion analysis. F, father; M, mother; Mk, 100 bp marker; P, patient. The arrowhead indicates the 500 bp marker. In the left panel, BseRI digested the 437 bp product of the father into two products of 147 and 290 bp. The 269C→T mutation in patient 1 and her mother removes the BseRI site, leaving the undigested 437 bp products present with the digested 147 and 290 bp products of the normal alleles, indicating that patient 1 and her mother are heterozygous for the 269C→T mutation. In the middle panel, Esp3I does not have a restriction site in the normal alleles of the patient's parents, leaving the 294 bp PCR product intact. The 937A→G mutation in patient 2 creates an Esp3I restriction site. Therefore, the 294 bp PCR product of the mutant allele of the patient is digested into two products of 139 and 155 bp, present along with the undigested 294 bp product of the normal allele, indicating that patient 2 is heterozygous for the 937A→G mutation. In the right panel, BstNI digested the 450 bp PCR product of the mother to two products of 138 and 312 bp. The 1690G→C mutation in patient 3 and her father removes the BstNI site, leaving the undigested 450 bp product present with the digested 138 and 312 bp products of the normal allele, indicating that patient 3 and her father are heterozygous for the 1690G→C mutation.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.