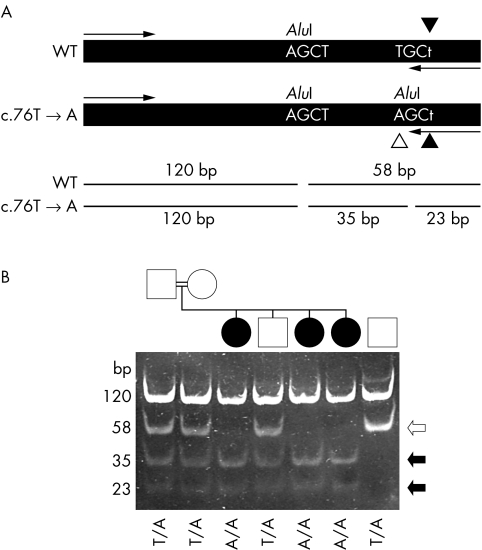
Figure 2 Analysis of the c.76T→A mutation. (A) Schematic diagram illustrating the approach taken to detect the mutation. A 236 bp fragment was amplified from genomic DNA is shown along with the primer annealing sites (arrows). The PCR using a mismatched reverse primer (c.79A→T change, indicated by filled arrowheads) was designed so that it introduced an AluI restriction enzyme digestion site into the PCR product only if the mutant (c.76T→A) gene sequence was present (identified by a white arrowhead). The restriction site was not introduced if the wild type sequence was present. An additional AluI site present in the amplified section from both normal and mutant alleles acted as a positive control for restriction enzyme digestion. Digestion of the amplified fragment with AluI at position 120 would produce 120 and 58 bp fragments. In the presence of the c.76T→A mutation, the 58 bp fragment would be cleaved into 35 and 23 bp fragments. (B) Analysis of samples from the French Canadian family LD28 (lanes 1 to 6) and a wild type control subject (lane 7). Samples in all lanes were digested with AluI. The genotype (c.76T→A mutation) of each individual is identified at the bottom of the gel picture. The sizes of each fragment are identified on the right side. Black and white arrows indicate fragments generated from the mutant and wild type alleles, respectively.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.