Abstract
Background
Characterisation of disease associated balanced chromosome rearrangements is a promising starting point in the search for candidate genes and regulatory elements.
Methods
We have identified and investigated three patients with limb abnormalities and breakpoints involving chromosome 2q31. Patient 1 with severe brachydactyly and syndactyly, mental retardation, hypoplasia of the cerebellum, scoliosis, and ectopic anus, carries a balanced t(2;10)(q31.1;q26.3) translocation. Patient 2, with translocation t(2;10)(q31.1;q23.33), has aplasia of the ulna, shortening of the radius, finger anomalies, and scoliosis. Patient 3 carries a pericentric inversion of chromosome 2, inv(2)(p15q31). Her phenotype is characterised by bilateral aplasia of the fibula and the radius, bilateral hypoplasia of the ulna, unossified carpal bones, and hypoplasia and dislocation of both tibiae.
Results
By fluorescence in situ hybridisation, we have mapped the breakpoints to intervals of approximately 170 kb or less. None of the three 2q31 breakpoints, which all mapped close to the HOXD cluster, disrupted any known genes.
Conclusions
Hoxd gene expression in the mouse is regulated by cis‐acting DNA elements acting over distances of several hundred kilobases. Moreover, Hoxd genes play an established role in bone development. It is therefore very likely that the three rearrangements disturb normal HOXD gene regulation by position effects.
Keywords: chromosome rearrangement, HOXD, limb malformation
The HOX genes code for a family of highly conserved transcription factors expressed during embryonic development. To date, mutations in four HOX genes, namely HOXA11, HOXA13, HOXD10, and HOXD13, have been found and all are associated with limb malformations. In particular, it has been shown that in two families a single nucleotide deletion within the second exon of HOXA11, which results in a frameshift and a premature stop codon, co‐segregates with proximal radial‐ulnar synostosis.1 Mutations in HOXA13 cause hand‐foot‐uterus syndrome, a rare dominantly inherited condition (OMIM 140000),2 and Guttmacher syndrome (OMIM 176305).3 Recently, a missense mutation in the HOXD10 gene has been described as the cause of isolated congenital vertical talus, also known as rocker‐bottom feet (OMIM 192950), and Charcot‐Marie‐Tooth disease (OMIM 118220).4 The first human limb malformation shown to be caused by HOXD13 polyalanine tract expansion mutations was synpolydactyly (SPD, OMIM 186000).5 Similar pathological polyalanine tract expansions in HOXD13 have subsequently been reported in other families with SPD.6,7,8 Intragenic frameshift deletions in HOXD13, predicted to result in truncated proteins, and also an acceptor splice site mutation and a missense mutation in exon 2 of HOXD13, cause an atypical form of SPD.9,10,11,12 Interestingly, a different missense mutation in the same exon has been found in a family with a dominantly inherited combination of brachydactyly and polydactyly.13
Large deletions involving chromosome 2q31.1 have been associated with minor digital anomalies,14,15 with major limb defects,15,16,17 or with a combination of severe limb and genital abnormalities.18 In contrast, a microdeletion at the 5′ end of the HOXD cluster that removes HOXD9 to HOXD13 and extends 85 kb upstream of HOXD13 results in an SPD phenotype.19
Another relevant point to consider is the presence of regulatory mutations affecting gene expression. Spitz et al described a balanced t(2;8) translocation that co‐segregated with mesomelic dysplasia and vertebral defects.20 The authors mapped the chromosome 2 breakpoint approximately 60 kb downstream from the HOXD cluster. Since no known gene is disrupted by the rearrangement, the authors speculated that the translocation affects regulatory sequences of the HOXD cluster, resulting in misregulation of HOXD gene expression.
In this paper, we report our findings on three unrelated patients with various skeletal malformations and balanced rearrangements involving chromosome band 2q31.
Methods
Subjects
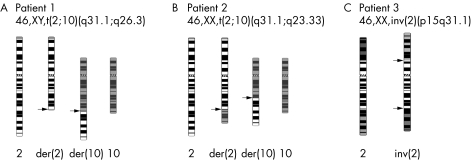
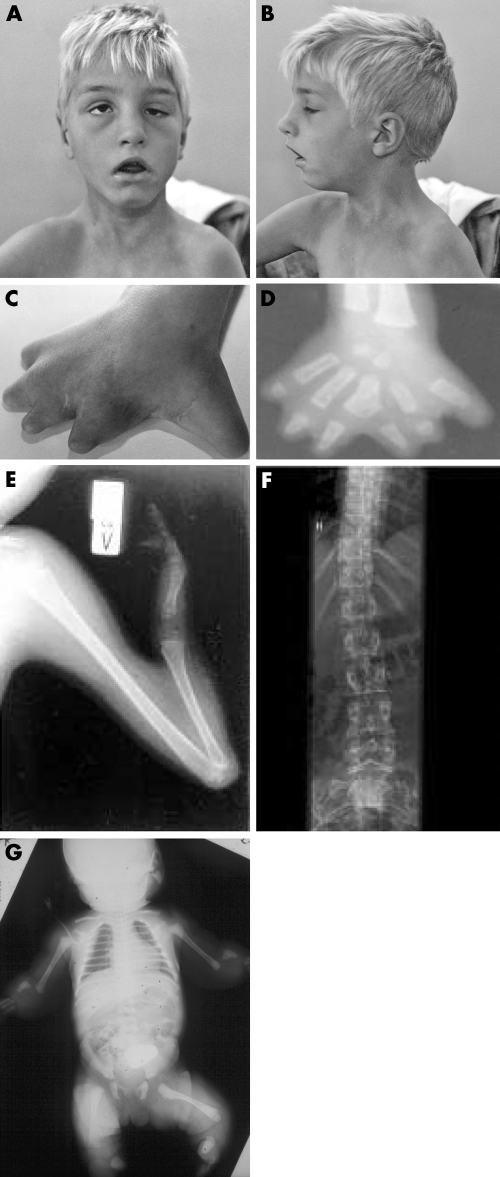
Proband 1 is a 13 year old boy carrying an apparently balanced chromosome rearrangement t(2;10)(q31.1;q26.3) (schematically depicted in fig 1A). He is the second child of healthy unrelated parents, born after a normal pregnancy and without fetal distress. Clinical examination after birth revealed severe malformation of the hands and feet as well as a dysmorphic face. The hands were short and had six fingers, and absence of the distal phalanges including the nails was noted. The feet showed complete absence of toes II–V, and a rudimentary first toe with a missing distal phalanx. x Rays of the hands showed five short metacarpals and six digits, each consisting only of a single phalanx. Metacarpal III was bifurcated at its end, giving rise to two digits. The patient was not able to fully extend his elbows and knees due to contractions. Bilateral inguinal hernias were noted and surgically corrected. x Rays of the thorax showed hypoplasia of the medial ends of both clavicles. Ultrasound of the skull revealed hypoplasia of the cerebellum. Further clinical examinations demonstrated developmental delay, deep set eyes (left more than right), a progressive scoliosis, narrow shoulders, ataxia, coxa valga, and short stature (110 cm at age 6, <3rd percentile) (fig 2A–D). The hands of the patient were surgical corrected and the polydactylous digits removed. The patient has developed well and is currently attending a special school.
Figure 1 Karyotypes of the three patients with apparently balanced chromosome rearrangements and limb malformations included in this study, and ideograms of the rearranged chromosomes. Arrows point to the breakpoints on der(2), der(10), and inv(2) chromosomes.
Figure 2 (A,B) Pictures of patient 1 at the age of 10. (C) Photo of the right hand of patient 1. (D) x Ray of the right hand of patient 1. Note the shortened metacarpals, webbing, duplications, and shortening of digits; (E) x Ray of the left arm of patient 2 at the age of 10 demonstrating ulnar aplasia, shortened radius, and absent 3rd to 5th rays. Furthermore, there is hypoplasia of digits 1 and 2. (F) x Ray of the spine of patient 2 at the age of 23 revealing a slight dextro‐convex scoliosis. (G) Babygram of proband 3. Note severe defects in distal limbs.
Proband 2 is a 23 year old female carrying a balanced de novo translocation t(2;10)(q31.1;q23.33) (fig 1B). She is the first child of healthy parents of Icelandic origin, born at term after an uncomplicated pregnancy. Except for her extremity malformation, her motor, mental, and social development were normal. She has an almost symmetrical limb phenotype consisting of ulnar aplasia, radial shortening, and absence of 3rd to 5th rays (fig 2E). There is hypoplasia of digits 1 and 2 where one epiphysial disc is shared by the two proximal phalangeal bones causing articulation between the fingers. Only one carpal bone is present. Humerus and lower limbs are normal. A slight dextro‐convex scoliosis was found at the age of 23 (fig 2F). Her mammary glands are normal.
Proband 3 is a newborn girl with a balanced de novo pericentric inversion inv(2)(p15q31.1). She is the first child of unrelated parents, a 30 year old mother and a 42 year old father of German origin. She was born after an uncomplicated pregnancy in the 40th week of gestation (birth weight 3700 g, length 49 cm, head circumference 37.5 cm). There was no history of any medication or illness during pregnancy. The first radiological examination after birth revealed bilateral aplasia of the fibula and the radius, bilateral hypoplasia of the ulna, unossified carpal bones, and hypoplasia and dislocation of both tibiae.
Breakpoint mapping by fluorescence in situ hybridisation (FISH)
YAC clones from the regions of interest were used for the initial mapping of the breakpoints, followed by fine mapping with smaller clones. FISH was carried out according to standard procedures. Genomic clones were labelled with digoxigenin‐dUTP or biotin‐dUTP and were hybridised to patient metaphase chromosomes. Signals were detected either by anti‐digoxigenin, FITC, or Cy3 conjugated avidin and were visualised by fluorescence microscopy with a CCD camera (Sensys, Photometrics, Tucson, AZ) and an image analysis program (IPLAB spectrum, Vysis, Downers Grove, IL). The spotted human chromosome 2‐specific cosmid library (Livermore) was hybridised with a pool of PCR products selected from the breakpoint region of patient 1. Positive cosmids were obtained as bacterial stocks from the Deutsche Ressourcenzentrum für Genomforschung (RZPD). DNAs were isolated with the Miniprep Kit (Qiagen, Hilden, Germany), and their ends were sequenced with T7 and T3 universal primers.
Southern blotting and breakpoint cloning
Genomic DNAs isolated from patient 1 and control cell lines were digested with BamHI, BglII, HindIII, or PstI restriction enzymes, separated in 1% agarose gels, transferred onto nylon membranes (Roth, Karlsruhe, Germany), and hybridised with [α32P]‐dCTP labelled PCR products.
Probes for Southern blot hybridisation were amplified using the following primer sets: 538A12_49750for (5′ GCT TCC CAT TGC AGG TGT AAA 3′) and 538A12_49750rev (5′ ATT ACT GGT CAT CAA TAT CTA GC 3′), 538A12_79400for (5′ AAC TCA ACA TAAACT TTT CCA AAG 3′) and 538A12_79400rev (5′ GAA TGT AAA ATA TAG ACA TTT GAC ATT G 3′) (positions 104665–105358 and 110085–110642 of clone RP11‐538A12, GenBank accession no. AC016761).
Breakpoint cloning was performed using adaptor ligated PCR as described elsewhere.21EcoRI digested patient DNA was PCR amplified with a set of nested primers, AP1 (5′ GGA TCC TAA TAC GAC TCA CTA TAG GGC 3′) with 84364for (5′ CAG ATT GTG ATT AGA TCA GGA G 3′) and AP2 (5′ TAT AGG GCT CGA GCG GC 3′) with 84715for (5′ GAC TTA AAA TTG CAG CGT GTG TTT C 3′) for der(10), and AP1 with 85206rev1 (5′ GTG TAT CTA TCT GAG CTC CAT G 3′) and AP2 with 85163rev2 (5′ TTC AGC CTT AAG TCA AAA TGT TGG 3′) for der(2). Amplified fragments were isolated from 1% agarose gels, subcloned into pGEM‐T Easy vector and sequenced using M13 universal primers.
Screening for HOXD13 mutation in patient 1
Mutation analysis of the HOXD13 gene in patient 1 included all coding exons and splice sites. Primers used for PCR amplifications are available online as supplementary data available at http://www.jmedgenet.com/supplemental.
Computational analysis of the breakpoint regions
Breakpoint regions were analysed in silico with the NIX program to identify known or predicted genes and ESTs. All putative ESTs were screened against entries in GenBank and TIGR databases.
Results
Mapping of the chromosome 2q31 breakpoints by FISH
FISH performed on patients' metaphase chromosomes showed that for patient 1, BAC clone RP11‐514D19 (GenBank accession number AC016915), which contains EVX2 and HOXD8‐13, showed signals on chromosome 2 and on der(10), indicating that the breakpoint lies outside this region (data not shown). Further hybridisation of a series of BAC clones selected from the region proximal to EVX2 led to the identification of the breakpoint‐spanning clone RP11‐538A12 (GenBank accession number AC016761) (fig 3A,D). To map the breakpoint more precisely, PCR products generated with primers selected from clone RP11‐538A12 were used for screening a chromosome 2‐specific cosmid library. Positive clones were hybridised to the patient chromosomes, and clone LLNLc128F0946 showed a signal on der(10), whereas clone LLNLc128A0237, with a partially overlapping sequence, showed a signal on der(2) (fig 3D).
Figure 3 FISH mapping of the 2q31 breakpoints in the patients. (A) Hybridisation of the breakpoint spanning clone RP11‐538A12 to patient 1 chromosomes; arrows indicate signals on chromosome 2, der(2), and der(10). (B) FISH results for patient 2 with RP11‐25L17, the breakpoint spanning BAC; arrows indicate signals on the normal and the derivative chromosomes. (C) Metaphase showing the breakpoint spanning YAC 888B4 in patient 3 with signals on the normal chromosome 2 and split signals on the inverted chromosome 2. (D) Schematic map of the 2q31.1 band showing the location of the three breakpoints in the vicinity of the HOXD complex together with breakpoint‐spanning BAC and cosmid clones; known genes from this region are depicted schematically. Approximate distances from the breakpoints to the HOXD13 gene are indicated by double headed arrows (not drawn to scale).
For patient 2, YAC 935E10 (with marker D2S138 and located at 192 cM) was found to span the 2q31 breakpoint (data not shown). Thus, the breakpoint of patient 2 was different from and more distal on chromosome 2q than the breakpoint of patient 1. This was confirmed by the FISH results obtained with BAC clone RP11‐387A1 (GenBank accession number AC009336), containing EVX2 and the complete HOXD cluster, which gave signals on chromosome 2 and on der(2) (data not shown). Hybridisation of BAC clone RP11‐25L17 (GenBank accession number AC096657) showed split signals on der(2) and der(10), indicating that this clone spans the breakpoint (fig 3B,D). Furthermore, BAC clones RP11‐1085F24 and RP11‐28M17 were also found to be breakpoint spanning (data not shown). All three breakpoint‐spanning BACs have overlapping sequences, with a common region of approximately 22 kb. Therefore, it is most likely that the breakpoint is located within this segment.
For patient 3, YAC clone 888B4 (with markers D2S148 and D2S2173 and located at 190 cM) was found to span the chromosome 2q31 breakpoint (fig 3C), indicating that this breakpoint is different from the two other breakpoints and lies in the region between the HOXD cluster and the breakpoint of patient 2. Further FISH studies showed that BAC clone RP11‐724O12 (GenBank accession number AC079803) spans the breakpoint (data not shown).
In conclusion, FISH mapping revealed that the chromosome 2q31 breakpoints investigated in the three patients with limb malformations lie outside the HOXD cluster: in patient 1 approximately 390 kb centromeric to HOXD13, in patient 2 approximately 1050 kb telomeric to HOXD13, and in patient 3 approximately 590 kb telomeric to HOXD13.
A similar approach was used to map the breakpoints on chromosome 10. In patient 1, BAC RP11‐300B2 (GenBank accession number AL355531) was breakpoint spanning. In patient 2, we were able to narrow the region to approximately 28 kb, between the clones RP11‐81C11 and RP11‐702G14 and clone RP11‐348J12 (GenBank accession number AL358613) (data not shown).
For patient 3, the breakpoint on the short arm of chromosome 2 was narrowed to a region of approximately 170 kb between clones RP11‐351H12 (GenBank accession numbers AQ529149 and AQ529148) and RP11‐355A23 (GenBank accession number AC009490) (data not shown).
Cloning of the breakpoints of patient 1
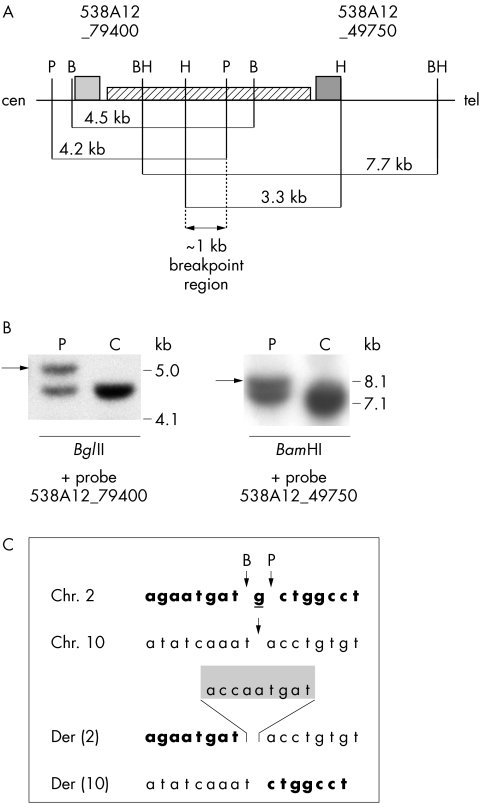
In order to localise precisely the chromosome 2 breakpoint of patient 1, we performed Southern blot hybridisation of patient and control DNAs digested with appropriate restriction enzymes. Since there is an approximately 4.5 kb long LINE repeat in the region of interest, we were very restricted with respect to designing probes. The two probes designed from sequences close to the repeat were most informative. Hybridisation of the more telomeric probe (538A12_49750) allowed observation of aberrant restriction fragments in patient DNA digested with BamHI, whereas the more centromeric probe (538A12_79400) showed aberrant restriction fragments in patient DNA cut with BglII (fig 4A,B). Mapping of the breakpoint on chromosome 10 gave us additional information about restriction sites on der(2) and der(10) chromosomes, and we found that EcoRI should be useful for our further analyses. In order to clone the breakpoint of chromosome 2, we ligated adaptors to EcoRI digested patient DNA. Subsequent PCR reactions gave rise to two products, approximately 500 bp long for der(10) and approximately 1.1 kb long for der(2), which were subcloned and sequenced. The breakpoint on chromosome 2 is located between positions 107 910 bp and 107 912 bp of clone RP11‐538A12. The nucleotide G at position 107 911 bp is deleted; moreover, on der(2), a 9 bp insertion is present between original chromosome 2 and chromosome 10 sequences (fig 4C).
Figure 4 (A) Schematic representation of the breakpoint region on BAC RP11‐538A12 and the localisation of probes 538A12_ 79400 and 538A12_ 49750 (shown as grey boxes) used for Southern blot hybridisation. The positions and sizes of normal restriction fragments are indicated and enzymes are marked as follows: B, BglII; BH, BamHI; H, HindIII; P, PstI; the striped box between the two probes indicates the LINE repeat. (B) Southern blot analysis with probe 538A12_ 49750; DNA from patient 1 (P) and a control (C) were digested with HindIII or BamHI restriction enzymes; the presence of additional bands in the patient compared to the control (arrows) indicates that the breakpoint is located within the HindIII and BamHI restriction fragments. (C) Chromosome 2, 10, der(2), and der(10) sequences in patient 1. Chromosome 2‐derived sequences are shown in bold; the underlined guanine residue from chromosome 2 is missing in both derivative chromosomes. The 9 bp insertion of unknown origin within der(2) (shaded box) separates chromosome 2‐ and chromosome 10‐derived sequences.
HOXD13 is not mutated in patient 1
The limb malformations of patient 1 resemble those present in patients with SPD and a mutation in HOXD13. Our mutation analysis of HOXD13 showed normal results. Hence, the phenotype is most likely caused by the translocation.
Computational analysis of the breakpoint regions
In silico analysis showed that there is no known gene on the chromosome 2q31 breakpoint‐spanning BAC (RP11‐538A12) in patient 1. For patient 2, we identified no known gene in the 22 kb long breakpoint interval; the closest gene (hnRNPA3) is located approximately 45 kb telomeric to the breakpoint region. For patient 3, sequence analysis of the breakpoint‐spanning BAC (RP11‐724O12) revealed the presence of three short unspliced ESTs, all derived from either cancer cells or stomach tissues, but none of these ESTs belongs to any known gene.
Our analysis of the chromosome 10 breakpoint region of patient 1 revealed that this breakpoint disrupts the MGMT gene, which encodes methylguanine‐DNA methyltransferase, an enzyme involved in DNA repair processes. MGMT is expressed from the intact chromosome 10 in the patient cell line (data not shown); thus, the breakpoint on chromosome 10 may play no role in the phenotype.
The breakpoint on chromosome 10 in patient 2 was mapped within a 28 kb region of the breakpoint‐spanning BAC clone RP11‐348J12. This region contains three genes: SEC15L1 and two cytochrome P450 genes, CYP26A1 and CYP26C1. The product of the human SEC15L1 gene is highly similar to the yeast protein SEC15, which is one of the components of a multiprotein complex required for exocytosis. To date, there is no indication that SEC15L1 plays a role in limb development. CYP26 family members of the cytochrome P450 catabolise and contribute to the control of retinoic acid, an important regulator of gene expression during embryonic development.22,23 The Cyp26a1−/− knock‐out mice show caudal truncation, vertebrae transformation, and hindbrain mispatterning. Moreover, in these mice the hindlimbs are fused, whereas no abnormalities of the upper limb structures are visible.24 The absence of lower limb malformations in patient 2 suggests that CYP26A1 is not involved in the observed upper limb malformations.
The breakpoint region on the short arm of chromosome 2 in patient 3 has been mapped to an interval of approximately 170 kb. This region contains no known gene.
Discussion
In the present study we have investigated three unrelated patients with severe limb malformations and apparently balanced chromosome rearrangements involving chromosome 2q31 and located close to the HOXD complex. It is noteworthy that computational analysis of each breakpoint region in 2q31 provided no indication that any known genes are directly truncated by the breakpoints. However, the patients show phenotypes characteristic for HOXD mutations which strongly suggests that the limb abnormalities are the result of disturbed HOXD expression through the chromosomal breakpoints rather than a coincidence of affected limb development by other mechanisms.
In patient 1, the phenotype resembles that of patients with SPD. The most common mutation causing SPD is an expansion of the imperfect trinucleotide repeat coding for alanine in the HOXD13 gene. Sequence analysis of the HOXD13 genes in patient 1 showed a normal result, which suggests that the phenotype is instead caused by effects of the translocation. It has been proposed that SPD is not only caused by a loss of function of the HOXD13 gene but also by a combined loss of function of several HOXD genes. This conclusion has been drawn from experiments done in the mouse. Hoxd13 homozygous knock‐out mice show a fairly mild phenotype25,26 which resembles neither that of patients with SPD nor that of spdh mice.27 Interestingly, mice with a deletion encompassing Hoxd11, Hoxd12, and Hoxd13 show a much more severe limb phenotype, comparable to that of the homozygous spdh mouse.28 Results obtained by several groups indicate that the posterior genes, Hoxa13, Hoxd11, Hoxd12, and Hoxd13, contribute significantly to digit development, and that these genes control this process in a dose‐dependent manner, whereas Hoxa11 and Evx2, the latter located at the centromeric end of the Hoxd complex, showed only a minor contribution to digit morphogenesis.29 We propose, therefore, that in patient 1 the translocation has an impact on the regulation of HOXD expression, which results in the HOXD loss of function phenotype. Aberrant gene expression caused by a change of the gene's chromosomal environment, referred to as a position effect, can be the result of various mechanisms, one of which is the separation of regulatory elements from their respective transcription unit, leading to misexpression (reviewed by Kleinjan and van Heyningen30,31). Intensive studies in the mouse led to the hypothesis that Hoxd10, Hoxd11, Hoxd12, and Hoxd13 genes, which show very similar expression domains in presumptive digits,32 are under the control of the same digit enhancer that controls their spatial and temporal expression in developing limbs. Furthermore, it has been suggested that this digit enhancer is located centromeric to the Hoxd complex.33 Many attempts have been made to find this enhancer sequence,34,35 but its precise location remains unknown. Introduction of a human PAC clone covering almost the entire HOXD cluster and extending approximately 40 kb centromeric to HOXD13 could not rescue the phenotype in mice which were triple mutants for Hoxd11/Hoxd12/Hoxd13, suggesting that the enhancer is located even further away from the cluster.36 More recently, a fragment of human DNA from the region more centromeric to the HOXD cluster has been shown to contain transcriptional enhancer activity similar to that controlling both Hoxd and Evx2 genes.37 This DNA segment is part of human BAC clone RP11‐504O20 (GenBank accession number AC016751), which partially overlaps with BAC 538A12, the clone spanning the breakpoint in patient 1. Spitz et al37 have also analysed BAC 538A12 but could not show any enhancer activity specific for posterior HOXD genes. It is thus very likely that the presumed digit enhancer lies close to the breakpoint in patient 1; however, it seems to be neither disrupted nor separated from the HOXD cluster by the breakpoint.
In patients 2 and 3, the 2q31 breakpoints are located on the telomeric side of the HOXD cluster, and both patients show malformations of more proximal parts of the limbs. In patient 2, the ulnae and the posterior digits in the hands are absent while the radii are hypoplastic. Patient 3 has no radii, ulnae, or fibulae, and shows hypoplastic tibiae. Improper development of the lower arm and lower legs has been described in mice lacking either Hoxd11 or Hoxa11 genes, as well as in the double mutants.38,39,40,41,42 Additionally, in Hoxd11 knock‐out mice, metacarpals, phalanges, and wrist bones are affected.39,40 Compound mutants for Hoxa10 and Hoxd11, as well as Hoxa10 and Hoxd10 double knock outs, showed more severe phenotypes, suggesting that, as for digit formation, zeugopod development is dependent on the correct expression of several posterior Hoxd genes.43,44 In addition to the knock‐out mice, a regulatory mutation in the mouse has been described. Ulnaless is a semidominant x ray induced mutation, which changes the level of posterior Hoxd expression in limb buds.33,45 The phenotype is very severe, with affected zeugopods and almost complete absence of ulnae, which resembles the phenotypes of patients 2 and 3. It has recently been shown that an inversion occurred on chromosome 2 in the ulnaless mouse, near the Hoxd cluster. The size of the inversion is approximately 770 kb, with one breakpoint centromeric to the Hoxd cluster disrupting the Lnp gene, and the other breakpoint, which probably does not truncate any gene, telomeric to the Hoxd cluster.37 Although the authors highlighted the relevance of the centromeric breakpoint for the ulnaless phenotype, our results for patients 2 and 3, together with a recent hypothesis about the early limb control region present on the telomeric site of the Hoxd cluster in mouse,46 suggest that the ulnaless breakpoint telomeric to Hoxd may also play a role in the observed zeugopod malformations. In line with this is the translocation t(2;8) described by Spitz et al,20 which is also associated with abnormal zeugopod development. In this translocation, the chromosome 2 breakpoint is located closer to the HOXD cluster than the breakpoint in patient 3 described in the present study. Similarly to the patients described in this study, however, no obvious gene was disrupted by the rearrangement. These findings indicate that sequences telomeric to the HOXD genes also play an important role in the regulation of the entire cluster. In light of this, it is likely that the limb phenotypes in patients 2 and 3 are the result of aberrant HOXD expression through the breakpoints telomeric to the HOXD cluster.
Other position effect mechanisms can also lead to gene silencing, either by insertion of a gene into a heterochromatic region or by removal of a long range insulator, which normally allows spreading of heterochromatin over long distances. However, it is rather unlikely that one of these mechanisms has caused misregulation of HOXD genes in patient 1, since the whole HOXD cluster and all currently known regulatory elements have been translocated into a transcriptionally active region at 10q26 containing the MGMT gene. Similarly, in patient 2, the breakpoint at 10q23.33 lies within a region containing several genes. Therefore, the phenotypes of these patients are more likely caused by translocation mediated disturbances in other mechanisms; for instance, the rearrangements might have brought the HOXD cluster into the vicinity of regulatory elements present near the second breakpoint. Alternatively, another transcription unit from the non‐HOXD breakpoint regions could compete with the HOXD genes for the digit enhancer or for the early limb control region, resulting in misregulation of the HOXD genes. Both hypotheses are interesting, however given the present state of knowledge, they are only speculative. At the moment, nothing is known about regulatory elements of MGMT or other genes in the vicinity of the breakpoint at 10q26 (EBF3 and TXNL2 are located approximately 330 kb and 630 kb telomeric to the chromosome 10 breakpoint in patient 1, and MKI67 is located approximately 1.4 Mb centromeric to this breakpoint), which could theoretically influence expression of the translocated HOXD cluster. Moreover, nothing is currently known about the presence of putative cis‐acting regulators in the breakpoint regions at 10q23 and 2p15 in patients 2 and 3.
Expression of posterior HOXD genes in limb buds is most likely regulated by the interplay between cis‐acting elements and trans‐acting factors. To date, two groups of genes, namely Polycomb (PcG) and trithorax (trxG) have been implicated in maintenance of the active or silent state of Hox genes in Drosophila (for review see Simon47) and mammals (reviewed in Schumacher and Magnuson48). Results from the last few years suggest that PcG genes may also play a role in controlling expression of posterior Hoxd genes in mouse limb buds by integrating both local and global regulatory mechanisms.49,50 However, it has been shown that binding of different trans‐acting regulators to DNA might be dependent on intact chromatin architecture.51,52 Since it has been proposed that the chromatin structure of any locus is determined by the combination of cis‐acting elements and the wider chromosomal and nuclear environment,31 it is plausible that chromosome rearrangements could influence chromatin architecture and thereby result in misregulation of genes. In fact, changes in chromatin structure have been proposed following insertion of some transgenes,53 or in the case of small deletions.54
Interestingly, changes in the global chromatin structure might also influence expression of other genes near the breakpoints at 2q31. The closest gene, located only 220 kb telomeric to the breakpoint in patient 1, is KIAA1715. This gene is transcribed from the strand opposite that of the HOXD cluster. Its mouse homologue, Lnp, shows the same expression pattern in limb buds and external genitalia as Hoxd13 and Evx2, which suggests that all three genes are under the control of the same regulatory sequences. In addition, Lnp is also expressed in the developing central nervous system in a highly similar pattern to that of Evx2, and it has a specific expression domain in the eyes, the heart, and the forebrain.37 The neural enhancer that may activate KIAA1715 is located in part within the same 40 kb region as the digit enhancer mentioned earlier; therefore, it is possible that both the limb and the neuronal expression domains of KIAA1715 have been affected by the translocation via a position effect. Since patient 1 has cognitive deficits in addition to limb abnormalities, it is tempting to link the central nervous system phenotype with disturbed expression of the KIAA1715 gene.
Although theoretically possible, it seems rather unlikely that the breakpoints on chromosome 10 and on the short arm of chromosome 2 cause the phenotypes of the patients reported here. The breakpoint at 10q26 in patient 1 disrupts the MGMT gene encoding the DNA‐repair enzyme methylguanine‐DNA methyltransferase; however, we observed expression of this gene from the intact chromosome 10 in patient lymphoblastoid cell lines. Moreover, MGMT−/− knock‐out mice are essentially normal,55 apart from being somewhat smaller than controls.56 For patient 2, three genes, SEC15L1, CYP26A1, and CYP26C1, reside on the chromosome 10‐derived breakpoint‐spanning clone RP11‐348J12; however, misexpression of none of these genes could explain her phenotype. In patient 3, the breakpoint in the short arm of chromosome 2 was mapped to an interval of approximately 170 kb. This interval and also its flanking DNA stretches, including 600 kb of 3′ sequence and 500 kb of 5′ sequence, harbour no known genes.
In summary, the data presented in this paper are plausible examples of regulatory mutations underlying severe developmental defects. They illustrate the complexity of HOXD cluster regulation and highlight the importance of the precise regulation of these genes for proper limb development.
Acknowledgements
We would like to thank Dr F Goodman for providing clones used for FISH in the initial phase of breakpoint mapping in patients 1 and 2. We also thank Dr S Shoichet for critical reading of the manuscript.
Abbreviations
FISH - fluorescence in situ hybridisation
SPD - synpolydactyly
Footnotes
This work was supported by a grant from the Deutsche Forschungsgemeinschaft to VK and SM, by the German Human Genome Program (DHGP, grant number 01KW99087), and by the National Genome Research Network (NGFN, project number 01GR0105). AS is supported by the Danish Research Agency. The Wilhelm Johannsen Centre for Functional Genome Research was established by the Danish National Research Foundation.
Competing interests: none declared
References
- 1.Thompson A A, Nguyen L T. Amegakaryocytic thrombocytopenia and radio‐ulnar synostosis are associated with HOXA11 mutation. Nat Genet 200026397–398. [DOI] [PubMed] [Google Scholar]
- 2.Mortlock D P, Innis J W. Mutation of HOXA13 in hand‐foot‐genital syndrome. Nat Genet 199715179–180. [DOI] [PubMed] [Google Scholar]
- 3.Innis J W, Goodman F R, Bacchelli C, Williams T M, Mortlock D P, Sateesh P, Scambler P J, McKinnon W, Guttmacher A E. A HOXA13 allele with a missense mutation in the homeobox and a dinucleotide deletion in the promoter underlies Guttmacher syndrome. Hum Mutat 200219573–574. [DOI] [PubMed] [Google Scholar]
- 4.Shrimpton A E, Levinsohn E M, Yozawitz J M, Packard D S, Jr, Cady R B, Middleton F A, Persico A M, Hootnick D R. A HOX gene mutation in a family with isolated congenital vertical talus and Charcot‐Marie‐Tooth disease. Am J Hum Genet 20047592–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muragaki Y, Mundlos S, Upton J, Olsen B R. Altered growth and branching patterns in synpolydactyly caused by mutations in HOXD13. Science 1996272548–551. [DOI] [PubMed] [Google Scholar]
- 6.Akarsu A N, Stoilov I, Yilmaz E, Sayli B S, Sarfarazi M. Genomic structure of HOXD13 gene: a nine polyalanine duplication causes synpolydactyly in two unrelated families. Hum Mol Genet 19965945–952. [DOI] [PubMed] [Google Scholar]
- 7.Kjaer K W, Hedeboe J, Bugge M, Hansen C, Friis‐Henriksen K, Vestergaard M B, Tommerup N, Opitz J M. HOXD13 polyalanine tract expansion in classical synpolydactyly type Vordingborg. Am J Med Genet 2002110116–121. [DOI] [PubMed] [Google Scholar]
- 8.Goodman F R, Mundlos S, Muragaki Y, Donnai D, Giovannucci‐Uzielli M L, Lapi E, Majewski F, McGaughran J, McKeown C, Reardon W, Upton J, Winter R M, Olsen B R, Scambler P J. Synpolydactyly phenotypes correlate with size of expansions in HOXD13 polyalanine tract. Proc Natl Acad Sci U S A 1997947458–7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodman F, Giovannucci‐Uzielli M L, Hall C, Reardon W, Winter R, Scambler P. Deletions in HOXD13 segregate with an identical, novel foot malformation in two unrelated families. Am J Hum Genet 199863992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debeer P, Bacchelli C, Scambler P J, De Smet L, Fryns J P, Goodman F R. Severe digital abnormalities in a patient heterozygous for both a novel missense mutation in HOXD13 and a polyalanine tract expansion in HOXA13. J Med Genet 200239852–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calabrese O, Bigoni S, Gualandi F, Trabanelli C, Camera G, Calzolari E. A new mutation in HOXD13 homeodomain causes a novel human limb malformation by producing selective loss of function. Eur J Hum Genet 20008(Suppl 1)140 [Google Scholar]
- 12.Kan S H, Johnson D, Giele H, Wilkie A O. An acceptor splice site mutation in HOXD13 results in variable hand, but consistent foot malformations. Am J Med Genet 2003121A69–74. [DOI] [PubMed] [Google Scholar]
- 13.Caronia G, Goodman F R, McKeown C M, Scambler P J, Zappavigna V. An I47L substitution in the HOXD13 homeodomain causes a novel human limb malformation by producing a selective loss of function. Development 20031301701–1712. [DOI] [PubMed] [Google Scholar]
- 14.Slavotinek A, Schwarz C, Getty J F, Stecko O, Goodman F, Kingston H. Two cases with interstitial deletions of chromosome 2 and sex reversal in one. Am J Med Genet 19998675–81. [DOI] [PubMed] [Google Scholar]
- 15.Nixon J, Oldridge M, Wilkie A O, Smith K. Interstitial deletion of 2q associated with craniosynostosis, ocular coloboma, and limb abnormalities: cytogenetic and molecular investigation. Am J Med Genet 199770324–327. [PubMed] [Google Scholar]
- 16.Boles R G, Pober B R, Gibson L H, Willis C R, McGrath J, Roberts D J, Yang‐Feng T L. Deletion of chromosome 2q24–q31 causes characteristic digital anomalies: case report and review. Am J Med Genet 199555155–160. [DOI] [PubMed] [Google Scholar]
- 17.Goodman F R. Limb malformations and the human HOX genes. Am J Med Genet 2002112256–265. [DOI] [PubMed] [Google Scholar]
- 18.Del Campo M, Jones M C, Veraksa A N, Curry C J, Jones K L, Mascarello J T, Ali‐Kahn‐Catts Z, Drumheller T, McGinnis W. Monodactylous limbs and abnormal genitalia are associated with hemizygosity for the human 2q31 region that includes the HOXD cluster. Am J Hum Genet 199965104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman F R, Majewski F, Collins A L, Scambler P J. A 117‐kb microdeletion removing HOXD9‐HOXD13 and EVX2 causes synpolydactyly. Am J Hum Genet 200270547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spitz F, Montavon T, Monso‐Hinard C, Morris M, Ventruto M L, Antonarakis S, Ventruto V, Duboule D. A t(2;8) balanced translocation with breakpoints near the human HOXD complex causes mesomelic dysplasia and vertebral defects. Genomics 200279493–498. [DOI] [PubMed] [Google Scholar]
- 21.Siebert P D, Chenchik A, Kellogg D E, Lukyanov K A, Lukyanov S A. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 1995231087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taimi M, Helvig C, Wisniewski J, Ramshaw H, White J, Amad M, Korczak B, Petkovich M. A novel human cytochrome P450, CYP26C1, involved in metabolism of 9‐cis and all‐trans isomers of retinoic acid. J Biol Chem 200427977–85. [DOI] [PubMed] [Google Scholar]
- 23.White J A, Beckett‐Jones B, Guo Y D, Dilworth F J, Bonasoro J, Jones G, Petkovich M. cDNA cloning of human retinoic acid‐metabolizing enzyme (hP450RAI) identifies a novel family of cytochromes P450. J Biol Chem 199727218538–18541. [DOI] [PubMed] [Google Scholar]
- 24.Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H. The retinoic acid‐inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio‐posterior axis within the mouse embryo. Genes Dev 200115213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis A P, Capecchi M R. A mutational analysis of the 5′ HoxD genes: dissection of genetic interactions during limb development in the mouse. Development 19961221175–1185. [DOI] [PubMed] [Google Scholar]
- 26.Dolle P, Dierich A, LeMeur M, Schimmang T, Schuhbaur B, Chambon P, Duboule D. Disruption of the Hoxd‐13 gene induces localized heterochrony leading to mice with neotenic limbs. Cell 199375431–441. [DOI] [PubMed] [Google Scholar]
- 27.Johnson K R, Sweet H O, Donahue L R, Ward‐Bailey P, Bronson R T, Davisson M T. A new spontaneous mouse mutation of Hoxd13 with a polyalanine expansion and phenotype similar to human synpolydactyly. Hum Mol Genet 199871033–1038. [DOI] [PubMed] [Google Scholar]
- 28.Zakany J, Duboule D. Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature 199638469–71. [DOI] [PubMed] [Google Scholar]
- 29.Zakany J, Duboule D. Hox genes in digit development and evolution. Cell Tissue Res 199929619–25. [DOI] [PubMed] [Google Scholar]
- 30.Kleinjan D J, van Heyningen V. Position effect in human genetic disease. Hum Mol Genet 199871611–1618. [DOI] [PubMed] [Google Scholar]
- 31.Kleinjan D A, van Heyningen V. Long‐range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 2005768–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sordino P, Duboule D. A molecular approach to the evolution of vertebrate paired appendages. Trends Ecol Evol 199611114–119. [DOI] [PubMed] [Google Scholar]
- 33.Peichel C L, Prabhakaran B, Vogt T F. The mouse Ulnaless mutation deregulates posterior HoxD gene expression and alters appendicular patterning. Development 19971243481–3492. [DOI] [PubMed] [Google Scholar]
- 34.Kmita M, Fraudeau N, Herault Y, Duboule D. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature 2002420145–150. [DOI] [PubMed] [Google Scholar]
- 35.Kondo T, Duboule D. Breaking colinearity in the mouse HoxD complex. Cell 199997407–417. [DOI] [PubMed] [Google Scholar]
- 36.Spitz F, Gonzalez F, Peichel C, Vogt T F, Duboule D, Zakany J. Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes Dev 2001152209–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 2003113405–417. [DOI] [PubMed] [Google Scholar]
- 38.Small K M, Potter S S. Homeotic transformations and limb defects in Hox A11 mutant mice. Genes Dev 199372318–2328. [DOI] [PubMed] [Google Scholar]
- 39.Davis A P, Capecchi M R. Axial homeosis and appendicular skeleton defects in mice with a targeted disruption of hoxd‐11. Development 19941202187–2198. [DOI] [PubMed] [Google Scholar]
- 40.Favier B, Le Meur M, Chambon P, Dolle P. Axial skeleton homeosis and forelimb malformations in Hoxd‐11 mutant mice. Proc Natl Acad Sci U S A 199592310–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis A P, Witte D P, Hsieh‐Li H M, Potter S S, Capecchi M R. Absence of radius and ulna in mice lacking hoxa‐11 and hoxd‐11. Nature 1995375791–795. [DOI] [PubMed] [Google Scholar]
- 42.Boulet A M, Capecchi M R. Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development 2004131299–309. [DOI] [PubMed] [Google Scholar]
- 43.Wahba G M, Hostikka S L, Carpenter E M. The paralogous Hox genes Hoxa10 and Hoxd10 interact to pattern the mouse hindlimb peripheral nervous system and skeleton. Dev Biol 200123187–102. [DOI] [PubMed] [Google Scholar]
- 44.Favier B, Rijli F M, Fromental‐Ramain C, Fraulob V, Chambon P, Dolle P. Functional cooperation between the non‐paralogous genes Hoxa‐10 and Hoxd‐11 in the developing forelimb and axial skeleton. Development 1996122449–460. [DOI] [PubMed] [Google Scholar]
- 45.Herault Y, Fraudeau N, Zakany J, Duboule D. Ulnaless (Ul), a regulatory mutation inducing both loss‐of‐function and gain‐of‐function of posterior Hoxd genes. Development 19971243493–3500. [DOI] [PubMed] [Google Scholar]
- 46.Zakany J, Kmita M, Duboule D. A dual role for Hox genes in limb anterior‐posterior asymmetry. Science 20043041669–1672. [DOI] [PubMed] [Google Scholar]
- 47.Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol 19957376–385. [DOI] [PubMed] [Google Scholar]
- 48.Schumacher A, Magnuson T. Murine Polycomb‐ and trithorax‐group genes regulate homeotic pathways and beyond. Trends Genet 199713167–170. [PubMed] [Google Scholar]
- 49.Barna M, Hawe N, Niswander L, Pandolfi P P. Plzf regulates limb and axial skeletal patterning. Nat Genet 200025166–172. [DOI] [PubMed] [Google Scholar]
- 50.Barna M, Merghoub T, Costoya J A, Ruggero D, Branford M, Bergia A, Samori B, Pandolfi P P. Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev Cell 20023499–510. [DOI] [PubMed] [Google Scholar]
- 51.Nourani A, Utley R T, Allard S, Cote J. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J 2004232597–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kornberg R D, Lorch Y. Chromatin structure and transcription. Annu Rev Cell Biol 19928563–587. [DOI] [PubMed] [Google Scholar]
- 53.de Graaff W, Tomotsune D, Oosterveen T, Takihara Y, Koseki H, Deschamps J. Randomly inserted and targeted Hox/reporter fusions transcriptionally silenced in Polycomb mutants. Proc Natl Acad Sci U S A 200310013362–13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang G, Yang F, van Overveld P G, Vedanarayanan V, van der Maarel S, Ehrlich M. Testing the position‐effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum Mol Genet 2003122909–2921. [DOI] [PubMed] [Google Scholar]
- 55.Glassner B J, Weeda G, Allan J M, Broekhof J L, Carls N H, Donker I, Engelward B P, Hampson R J, Hersmus R, Hickman M J, Roth R B, Warren H B, Wu M M, Hoeijmakers J H, Samson L D. DNA repair methyltransferase (Mgmt) knockout mice are sensitive to the lethal effects of chemotherapeutic alkylating agents. Mutagenesis 199914339–347. [DOI] [PubMed] [Google Scholar]
- 56.Tsuzuki T, Sakumi K, Shiraishi A, Kawate H, Igarashi H, Iwakuma T, Tominaga Y, Zhang S, Shimizu S, Ishikawa T.et al Targeted disruption of the DNA repair methyltransferase gene renders mice hypersensitive to alkylating agent. Carcinogenesis 1996171215–1220. [DOI] [PubMed] [Google Scholar]