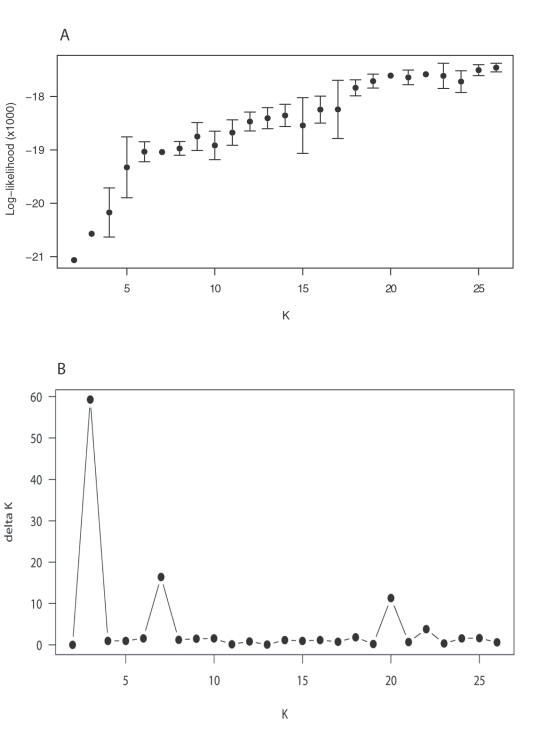
Figure 2. Results of the fully Bayesian model-based clustering method implemented in STRUCTURE v2.2 [21], which was used to infer the most likely number of populations (K) in the data set.
For this purpose, the program was run without population information under the admixture model (individuals may have mixed ancestry) and independent allele frequencies. Length of the burn-in was 100,000 and the number of MCMC replications after the burn-in was 1,000,000. Five independent chains were run for each K from K = 2 to K = 26. A) Ln P for each K for K = 2–26, B) ΔK for each K for K = 2–26 [52].