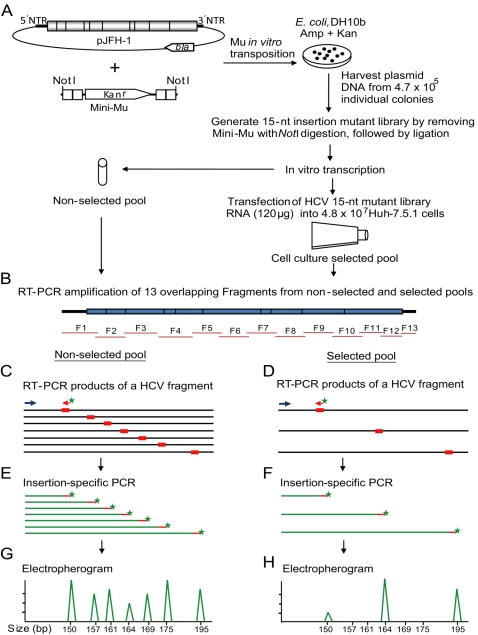
Figure 1. Schematic diagram depicting the various steps involved in Hepatitis C virus functional profiling.
(A) The plasmid carrying the JFH-1 HCV genome is subjected to in vitro mutagenesis by using the mini-Mu transposon, then selected in E. coli bacteria. The harvested mutant plasmids are subjected to NotI restriction enzyme digestion to remove the transposon body, followed by ligation resulting in the generation of a 15-nt insertion plasmid library. Subsequently, this mutant plasmid library is in vitro transcribed and used as a non-selected input pool for functional profiling analysis. The in vitro transcribed RNA library is delivered into Huh-7.5.1 cells for genetic selection. The total RNA harvested from the transfected cells (selected pool) as well as non-selected pool RNA are subjected to functional profiling analysis. (B) Followed by selection, the mutant HCV genome from the non-selected input pool and the selected pool are reverse-transcribed and 13 overlapping fragments (F1 to F13) are PCR amplified. (C, D) The purified PCR products from non-selected and selected pools are used as templates for a second PCR using one of the HCV fragment-specific primers (blue arrow) and a fluorescently labeled insertion-specific primer (red arrow with green star). (E, F) The fluorescently-labeled PCR products from input and selected pools are analyzed by a 96-capillary genotyper. The processed data are either visualized by electropherograms (G, H) or exported as a data file. The phenotype for each insertion is calculated by comparing the corresponding peak areas of selected and non-selected pools.