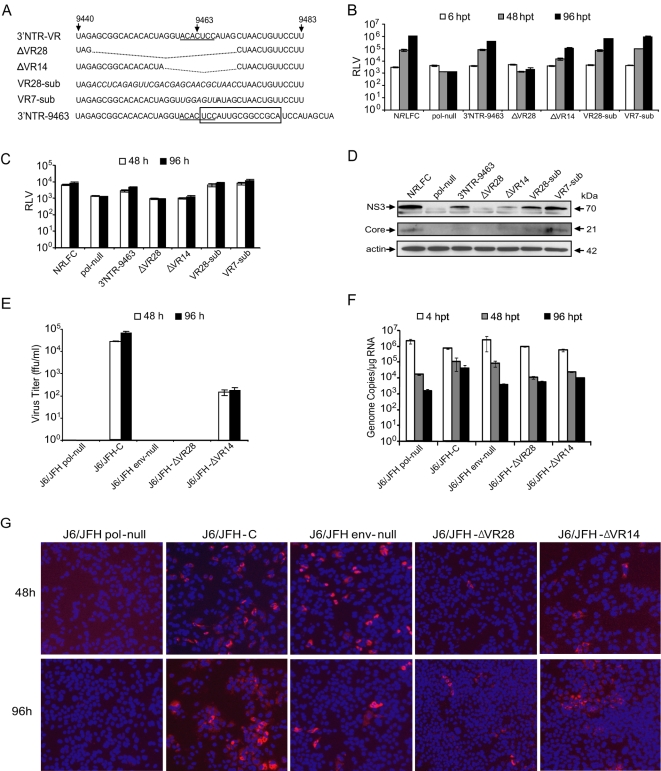
Figure 7. Analysis of 3′NTR Variable Region in HCV replication.
(A) Nucleotide sequence of JFH-1 3′NTR variable region and the mutations engineered in the individual mutant viruses are depicted. The genome position of the nucleotides is indicated. The sequence that is conserved across all the genotypes is underlined. The deleted polynucleotide regions are shown in dotted lines. The substituted heterologous polynucleotides are in italics. 15-nt insertion sequence is boxed. Due to space limitation, only a partial sequence is shown for 3′NTR-9463 mutant. (B) Analysis of viral genome replication of mutant reporter viruses. The mean Renilla luciferase values (RLV) with standard deviations are shown in the graph (log10 scale). (C) Measuring the production of infectious viral particles by mutant reporter viruses. The mutants deficient in production of infectious particles show only a background level of luciferase activity. (D) Western blotting analysis of viral proteins, NS3 and core, expression. (E) Comparison of J6/JFH-C mutant viral infectivity. The virus titer (ffu/ml) of cell-free supernatant collected at 48 and 96 hpt of J6/JFH-C based mutants' transfected cell culture was measured by infecting naïve Huh-7.5.1 cells. Mean values and standard deviations are shown in the graph. (F) Comparison of J6/JFH-C mutant viral genome replication. At 4, 48, and 96 hpt total cellular RNAs were harvested and subjected to RT-qPCR. The genome copy numbers per µg of RNA are presented. (G) Immunofluorescence assay. At 48 and 96 hpt the cells were fixed and stained for HCV core antigen. The cell nuclei were visualized by DAPI staining. For B, C, and D experimental details see Figure 6 legend.