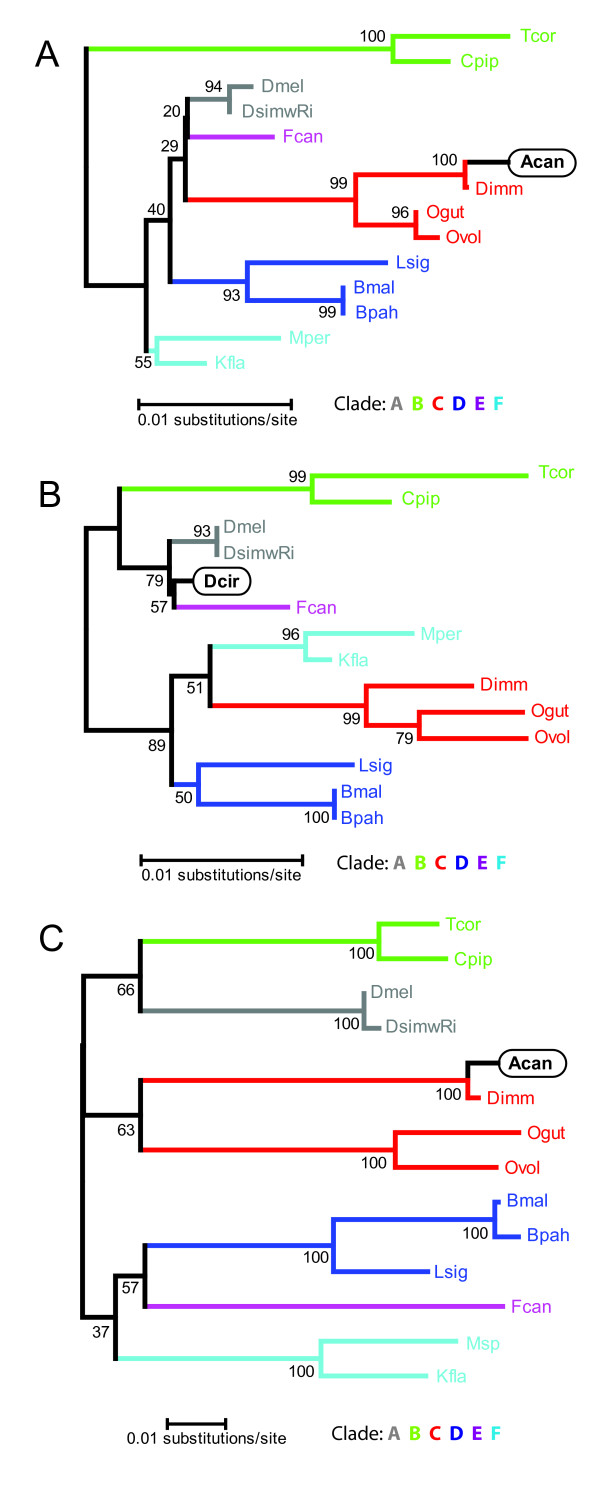
Figure 1.
Minimum evolution trees based on alignments of A) the Wolbachia16S (770 nucleotides) reported from A. cantonensis[GenBank:AY652762], B) the Wolbachia 16S (639 nucleotides) of D. circumlita c2 [GenBank:AY486072], and C) the Wolbachia ftsZ (431 nucleotides) reported from A. cantonensis [GenBank:DQ159068 ]. Sequences were aligned using ClustalX version 2.0.7 [14] using default parameters for slow/accurate alignment [Gap Opening:10, Gap Extend: 0.1, IUB DNA weight matrix]. After alignment, sequences were manually trimmed to the endpoints of the 16S sequences of the Wolbachia from A. cantonensis (see additional file 1: Wolbachia 16S multiple sequence alignment – A. cantonensis) and D. circumlita (see additional file 2: Wolbachia 16S multiple sequence alignment – D. circumlita), and to the endpoints of the ftsZ sequence of the Wolbachia from A. cantonensis (see additional file 3: Wolbachia ftsZ multiple sequence alignment). Phylogenetic trees were calculated using the Minimum Evolution method in MEGA4 [15]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Evolutionary distances were computed using the Maximum Composite Likelihood. The Minimum Evolution tree was searched using the Close-Neighbor-Interchange (CNI) algorithm at a search level of 1. The Neighbor-joining algorithm was used to generate the initial tree. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option).