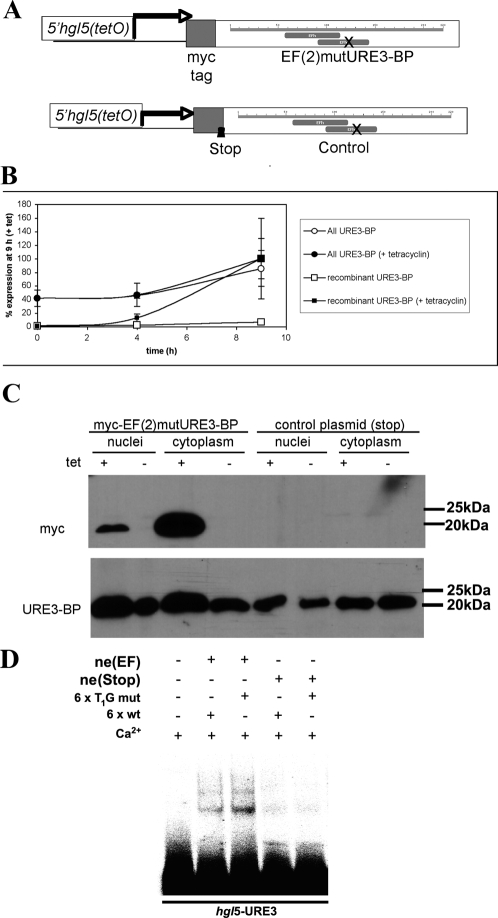
Figure 3. Inducible Overexpression of a Calcium-Insensitive Mutant of URE3-BP (EF(2)mutURE3-BP) in E. histolytica.
A recombinant version of URE3-BP was generated by mutating one of the two EF-hand motifs of URE3-BP (associated with the ability to bind calcium, EF(2)mutURE3-BP [10]) and by introducing an N terminal myc tag. As calcium inhibited DNA binding by URE3-BP, this generated a dominant positive mutant [10]. The recombinant protein was placed under the control of a tetracycline-inducible gene expression system of E. histolytica (previously described by Ramakrishnan et al. [27]). As a control, in a second construct the initial N terminal sequence of URE3-BP was replaced by the sequence CTTGTATTTAACAATAGCTAACATC, mutated bases underlined, which introduced stop codons into the two open reading frames at the N terminus. (A) Cartoon showing the salient features of the constructs. (B) qRT-PCR of un-induced and induced ameba transfected with the pEF((2))mutURE3-BP. Results are normalized to the levels of lgl1 and shown as a percentage of values of ameba induced for 9 h (y axis). Time after induction is shown on the X axis. (C) Western blot of nuclear and cytoplasmic extracts from tetracycline-induced and un-induced amebae probed with an antibody specific for the myc tag and therefore the recombinant protein (9E10), as well as with a monoclonal antibody to URE3-BP (4D6). (D) Calcium insensitive binding to URE3 DNA in extract prepared from EF(2)mutURE3-BP transformed trophozoites. EMSA performed with added calcium and radioactively labeled hgl5-URE3 double-stranded DNA. Other than the lane with probe alone, reactions included 2 μg of E. histolytica nuclear extract prepared from either induced trophozoites carrying EF(2)mutURE3-BP or as a control STOP- EF(2)mutURE3-BP. A six fold excess of either cold hgl5-URE3 (wt), or an oligonucleotide with a base pair change which substituted a G for a T at the first position and had no impact on URE3-BP specific band formation (T1G mut) were added as shown.