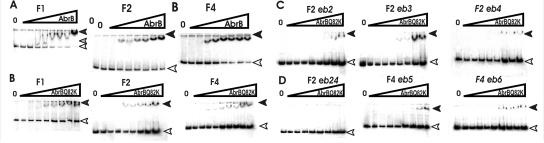
FIG. 4.
Gel retardation mobility shift assays in the presence of increasing concentrations of AbrB and AbrBQ82K. AbrB concentrations used were 0 (for lanes 0) and 0.25, 0.5, 0.75, 1, 1.25, 1.5, and 2 μM (from left to right for the lanes marked with gradients). (A) Gel shift analysis performed with DNA fragments of different lengths bound to AbrB. (B) Assay corresponding to that for panel A but performed with AbrBQ82K. Fragment F1 harbors the entire phyC promoter region with both AbrB binding regions, fragment F2 harbors region 1, and fragment F4 harbors region 2. (C and D) Effect of mutations eb2, eb3, eb4, and eb24 introduced into AbrB binding region 1 on the mobility of the F2 fragment and effect of mutations eb5 and eb6 within AbrB binding region 2 on the mobility of the F4 fragment. White arrowheads, free DNA; filled arrowheads, protein-DNA complex.